

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MLMASTTSAVPGHPSLPSLPSNSSQERPLDTRDPLLARAELALLSIVFVAVALSNGLVLAALARRGRRGHWAPIHVFIGHLCLADLAVALFQVLPQLAWKATDRFRGPDALCRAVKYLQMVGMYASSYMILAMTLDRHRAICRPMLAYRHGSGAHWNRPVLVAWAFSLLLSLPQLFIFAQRNVEGGSGVTDCWACFAEPWGRRTYVTWIALMVFVAPTLGIAACQVLIFREIHASLVPGPSERPGGRRRGRRTGSPGEGAHVSAAVAKTVRMTLVIVVVYVLCWAPFFLVQLWAAWDPEAPLEGAPFVLLMLLASLNSCTNPWIYASFSSSVSSELRSLLCCARGRTPPSLGPQDESCTTASSSLAKDTSS | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCSSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCC | |
| 91788888888998877888888888887878701999999999889899899899760311356887889611999999999999999998999999999294755348998999999999999999999999986999771087401107899999999999999999999999807888358985478830189724789999999999999999999999999999997635887420045665302455542045788888888777889999999993899999999996767531379999999999999989999999649999999999828776789987777788701267763304589 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MLMASTTSAVPGHPSLPSLPSNSSQERPLDTRDPLLARAELALLSIVFVAVALSNGLVLAALARRGRRGHWAPIHVFIGHLCLADLAVALFQVLPQLAWKATDRFRGPDALCRAVKYLQMVGMYASSYMILAMTLDRHRAICRPMLAYRHGSGAHWNRPVLVAWAFSLLLSLPQLFIFAQRNVEGGSGVTDCWACFAEPWGRRTYVTWIALMVFVAPTLGIAACQVLIFREIHASLVPGPSERPGGRRRGRRTGSPGEGAHVSAAVAKTVRMTLVIVVVYVLCWAPFFLVQLWAAWDPEAPLEGAPFVLLMLLASLNSCTNPWIYASFSSSVSSELRSLLCCARGRTPPSLGPQDESCTTASSSLAKDTSS | |
| 33230232222333323323333233333322222001010221120011013011000000011352343000000000002000000100010100000042220020000002001000000000000000132010000002023322321010000000000000000000000023155442101010101262021000001022313310110020001000101332343546444444444444444444323423120000000001000000211000000000134231220010000000020001000000000430040023002023434664444454333334233345438 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCSSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCC MLMASTTSAVPGHPSLPSLPSNSSQERPLDTRDPLLARAELALLSIVFVAVALSNGLVLAALARRGRRGHWAPIHVFIGHLCLADLAVALFQVLPQLAWKATDRFRGPDALCRAVKYLQMVGMYASSYMILAMTLDRHRAICRPMLAYRHGSGAHWNRPVLVAWAFSLLLSLPQLFIFAQRNVEGGSGVTDCWACFAEPWGRRTYVTWIALMVFVAPTLGIAACQVLIFREIHASLVPGPSERPGGRRRGRRTGSPGEGAHVSAAVAKTVRMTLVIVVVYVLCWAPFFLVQLWAAWDPEAPLEGAPFVLLMLLASLNSCTNPWIYASFSSSVSSELRSLLCCARGRTPPSLGPQDESCTTASSSLAKDTSS | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.24 | 0.24 | 0.80 | 2.76 | Download | ------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK--TATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL------------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.17 | 0.24 | 0.86 | 3.82 | Download | ---------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMR--NGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDKGSYLRICLLHPVQKFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLREGGGSGGDEAEKLFNQRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC----------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.24 | 0.24 | 0.80 | 3.62 | Download | ------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK--TATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV------------------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.17 | 0.20 | 0.96 | 1.55 | Download | -DLRDNETW-----WYNPSIIVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKS--LQTPANMFIINLAFSDFTFSLVNGFPLMISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLE--GVLCNCSFDYISRSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMA---KRL-N-AKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLQFDDKETDDAGESSDAAPSADAAQMKE | |||||||||||||||||||
| 5 | 4djh | 0.22 | 0.22 | 0.82 | 1.17 | Download | --------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMK--TATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE-DVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEMLRAIQKRTGTWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.24 | 0.24 | 0.80 | 3.04 | Download | --------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK--TATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL------------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.16 | 0.21 | 0.80 | 1.75 | Download | -------------------------------------VFIVLVAGSLSLVTIIGNILVMVSIKVN--RHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFS---NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLEGGNAAITKNRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC-IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------ | |||||||||||||||||||
| 8 | 4n6hA | 0.25 | 0.24 | 0.78 | 4.53 | Download | -------------------------------GSPALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKM--KTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD---GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.23 | 0.24 | 0.80 | 2.83 | Download | ------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK--TATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.22 | 0.22 | 0.78 | 3.56 | Download | ---------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMK--TATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQG---SIDCTLTFPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLK------------------SVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
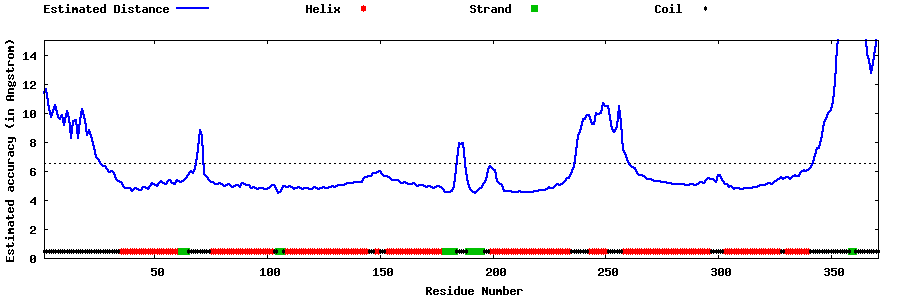
| Generated 3D models | Estimated local accuracy of models | ||
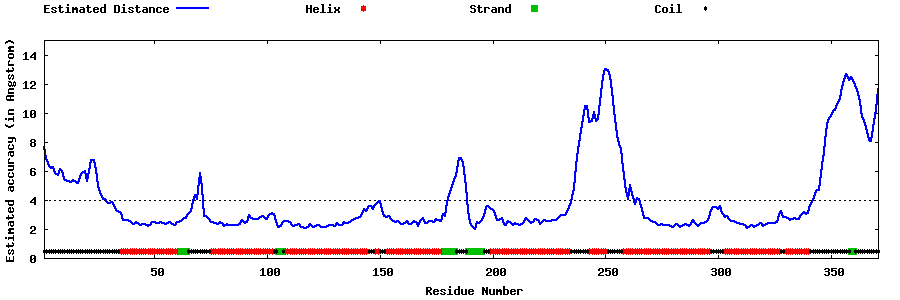 | |||
|
|
|||
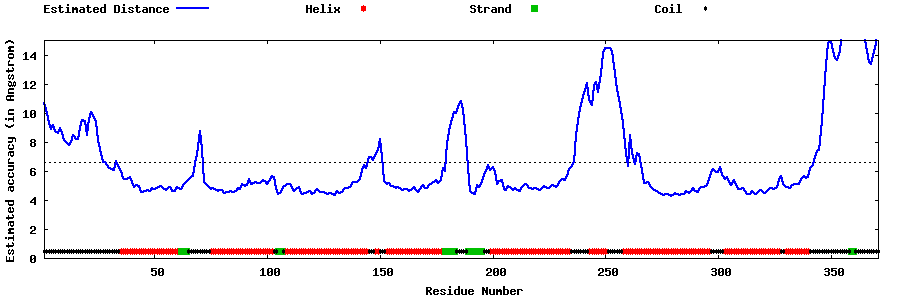 | |||
|
| |||
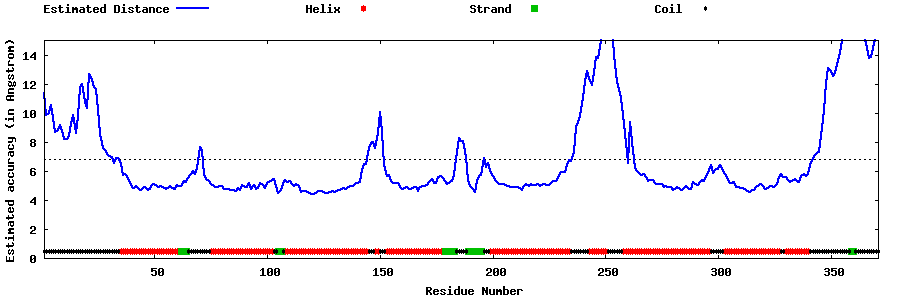 | |||
|
| |||
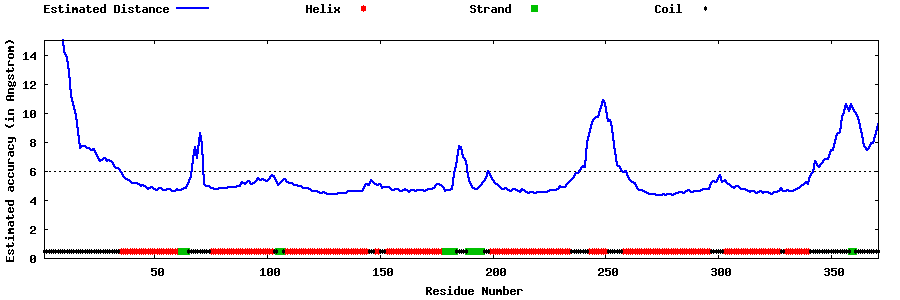 | |||
|
| |||
 | |||
|
| |||