

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| METLCLRASFWLALVGCVISDNPERYSTNLSNHVDDFTTFRGTELSFLVTTHQPTNLVLPSNGSMHNYCPQQTKITSAFKYINTVISCTIFIVGMVGNATLLRIIYQNKCMRNGPNALIASLALGDLIYVVIDLPINVFKLLAGRWPFDHNDFGVFLCKLFPFLQKSSVGITVLNLCALSVDRYRAVASWSRVQGIGIPLVTAIEIVSIWILSFILAIPEAIGFVMVPFEYRGEQHKTCMLNATSKFMEFYQDVKDWWLFGFYFCMPLVCTAIFYTLMTCEMLNRRNGSLRIALSEHLKQRREVAKTVFCLVVIFALCWFPLHLSRILKKTVYNEMDKNRCELLSFLLLMDYIGINLATMNSCINPIALYFVSKKFKNCFQSCLCCCCYQSKSLMTSVPMNGTSIQWKNHDQNNHNTDRSSHKDSMN | |
| CCHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHSSHHHHHHHHHHHHHHHHHSSSSSSSSCCCCSSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCC | |
| 9405866999998665533699612346887555666677777643354678887747788987666786666640289999999999999998688998779753179899799999999999999999998889999999705765787471222007999999999999999999999849897524322406126758511508999999999999998589996306996379986379428899999999999989799999999999999999971658997642467888861328888899999999998999999999999743378431539999999999999999999999999999819999999999961527988875677788887121247997889888877789999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| METLCLRASFWLALVGCVISDNPERYSTNLSNHVDDFTTFRGTELSFLVTTHQPTNLVLPSNGSMHNYCPQQTKITSAFKYINTVISCTIFIVGMVGNATLLRIIYQNKCMRNGPNALIASLALGDLIYVVIDLPINVFKLLAGRWPFDHNDFGVFLCKLFPFLQKSSVGITVLNLCALSVDRYRAVASWSRVQGIGIPLVTAIEIVSIWILSFILAIPEAIGFVMVPFEYRGEQHKTCMLNATSKFMEFYQDVKDWWLFGFYFCMPLVCTAIFYTLMTCEMLNRRNGSLRIALSEHLKQRREVAKTVFCLVVIFALCWFPLHLSRILKKTVYNEMDKNRCELLSFLLLMDYIGINLATMNSCINPIALYFVSKKFKNCFQSCLCCCCYQSKSLMTSVPMNGTSIQWKNHDQNNHNTDRSSHKDSMN | |
| 6311101000000001002023333222113332132232323322233333323222233233334222342413200100000011200100131120000000003402100000000002001010000000100210043022331100100010000001000000000000001303100000021233213200000000011100000000000010131414443100000001342341021000101133331310000020000000101323345444434533442110000000000021013121000100000023313433230100000011101000020000000000001431140024001012234454444343433324343454444434433444348 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHSSHHHHHHHHHHHHHHHHHSSSSSSSSCCCCSSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCC METLCLRASFWLALVGCVISDNPERYSTNLSNHVDDFTTFRGTELSFLVTTHQPTNLVLPSNGSMHNYCPQQTKITSAFKYINTVISCTIFIVGMVGNATLLRIIYQNKCMRNGPNALIASLALGDLIYVVIDLPINVFKLLAGRWPFDHNDFGVFLCKLFPFLQKSSVGITVLNLCALSVDRYRAVASWSRVQGIGIPLVTAIEIVSIWILSFILAIPEAIGFVMVPFEYRGEQHKTCMLNATSKFMEFYQDVKDWWLFGFYFCMPLVCTAIFYTLMTCEMLNRRNGSLRIALSEHLKQRREVAKTVFCLVVIFALCWFPLHLSRILKKTVYNEMDKNRCELLSFLLLMDYIGINLATMNSCINPIALYFVSKKFKNCFQSCLCCCCYQSKSLMTSVPMNGTSIQWKNHDQNNHNTDRSSHKDSMN | |||||||||||||||||||||||||
| 1 | 5glhA | 0.66 | 0.46 | 0.68 | 2.88 | Download | ------------------------------------------------------------------PPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYK-----NGPNILIASLALGDLLHIVIAIPINVYKLLAEDWP-----FGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWWTA----------VEIVLIWVVSVVLAVPEAIGFDIITMDYKGSYLRICLLHPVQKFMQFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKN---------DHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNRCELLSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSAL------------------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.66 | 0.49 | 0.74 | 3.76 | Download | ----------------------------------------------------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAEDW-----PFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDYKGSYLRICLLHPVQKFMQFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNRCELLSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC----------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.25 | 0.20 | 0.71 | 3.23 | Download | -------------------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWP-----FGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD----GAVVCMLQFPSP-SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD------PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.27 | 0.22 | 0.68 | 1.52 | Download | ---------------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWP-----FGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR--EDVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA------------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.26 | 0.22 | 0.68 | 1.17 | Download | ---------------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSW-----PFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE--DVDVIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNKTYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.22 | 0.23 | 0.86 | 3.42 | Download | NAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWP-----FGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD----GAVVCMLQFP-SPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG---SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRR------DPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.27 | 0.22 | 0.67 | 1.71 | Download | ------------------------------------------------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSW-----PFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVD--VIECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA----A--------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------------------------------ | |||||||||||||||||||
| 8 | 4n6hA | 0.26 | 0.20 | 0.71 | 4.21 | Download | --------------------------------------------------------------GS----PGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWP-----FGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD----GAVVCMLQFPSP-SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD------PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------- | |||||||||||||||||||
| 9 | 5glhA | 0.67 | 0.46 | 0.68 | 3.31 | Download | ------------------------------------------------------------------PPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIY-----KNGPNILIASLALGDLLHIVIAIPINVYKLLAEDWP-----FGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWW----------TAVEIVLIWVVSVVLAVPEAIGFDIITMDYKGSYLRICLLHPVTAFMQFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKN---------DHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNRCELLSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSAL------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.26 | 0.21 | 0.69 | 3.25 | Download | ----------------------------------------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWP-----FGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR----QGSIDCTLTFSHP-TWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS---KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
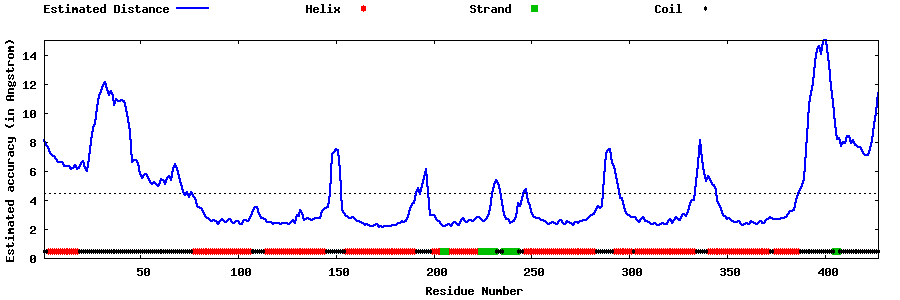 | |||
|
|
|||
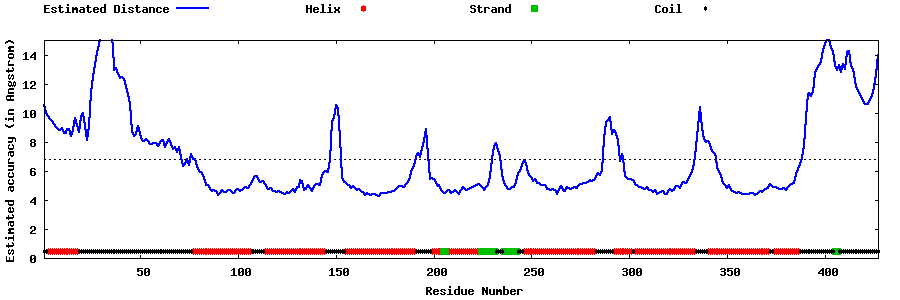 | |||
|
| |||
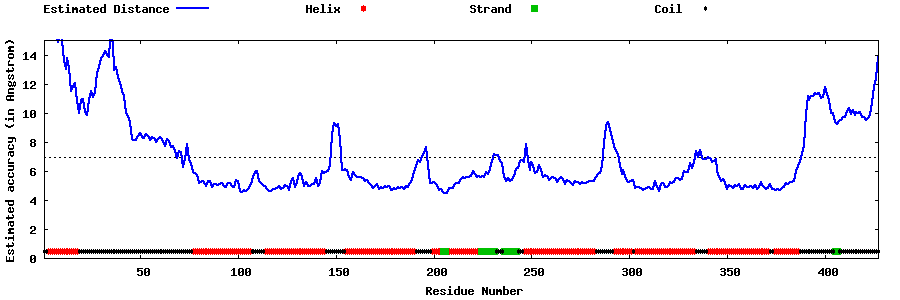 | |||
|
| |||
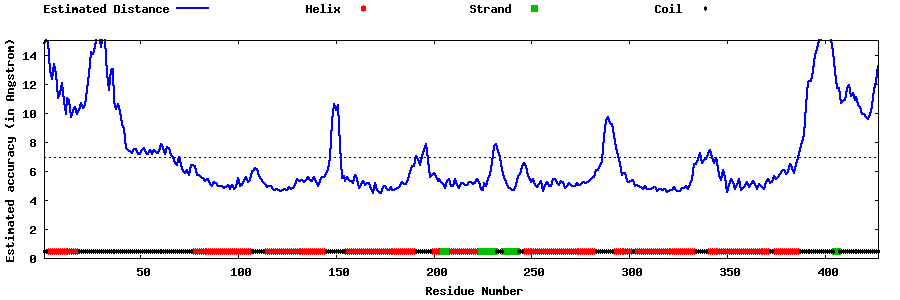 | |||
|
| |||
 | |||
|
| |||