

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MAPWPHENSSLAPWPDLPTLAPNTANTSGLPGVPWEAALAGALLALAVLATVGGNLLVIVAIAWTPRLQTMTNVFVTSLAAADLVMGLLVVPPAATLALTGHWPLGATGCELWTSVDVLCVTASIETLCALAVDRYLAVTNPLRYGALVTKRCARTAVVLVWVVSAAVSFAPIMSQWWRVGADAEAQRCHSNPRCCAFASNMPYVLLSSSVSFYLPLLVMLFVYARVFVVATRQLRLLRGELGRFPPEESPPAPSRSLAPAPVGTCAPPEGVPACGRRPARLLPLREHRALCTLGLIMGTFTLCWLPFFLANVLRALGGPSLVPGPAFLALNWLGYANSAFNPLIYCRSPDFRSAFRRLLCRCGRRLPPEPCAAARPALFPSGVPAARSSPAQPRLCQRLDGASWGVS | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 998998887777787777778777777788887499999999999999999998887167577016888614799999999999999999899999998270568758999999999999999999999999977773574224875253888888657999999999999998186445766666511357875565477289899999999999999999999999999999876402345643334544555432112345443222345543320004567778789876789999999958999999999983675684799999999999998475699818799999999997264678999876554666577887644568888763433678899999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MAPWPHENSSLAPWPDLPTLAPNTANTSGLPGVPWEAALAGALLALAVLATVGGNLLVIVAIAWTPRLQTMTNVFVTSLAAADLVMGLLVVPPAATLALTGHWPLGATGCELWTSVDVLCVTASIETLCALAVDRYLAVTNPLRYGALVTKRCARTAVVLVWVVSAAVSFAPIMSQWWRVGADAEAQRCHSNPRCCAFASNMPYVLLSSSVSFYLPLLVMLFVYARVFVVATRQLRLLRGELGRFPPEESPPAPSRSLAPAPVGTCAPPEGVPACGRRPARLLPLREHRALCTLGLIMGTFTLCWLPFFLANVLRALGGPSLVPGPAFLALNWLGYANSAFNPLIYCRSPDFRSAFRRLLCRCGRRLPPEPCAAARPALFPSGVPAARSSPAQPRLCQRLDGASWGVS | |
| 743233523333323423433333344342432302100000000200220231120000000113301100000000001000100000022000000243320131020000000010000001000000000000002003033312320000000000120121021000000133435443342344432020223310000000100310000002001200100231244243343434333223222322233333334333333233433321232322300310000000001001000000002000340200310000000201200110000000142003001200010233345443434333324443333433453343134266364348 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MAPWPHENSSLAPWPDLPTLAPNTANTSGLPGVPWEAALAGALLALAVLATVGGNLLVIVAIAWTPRLQTMTNVFVTSLAAADLVMGLLVVPPAATLALTGHWPLGATGCELWTSVDVLCVTASIETLCALAVDRYLAVTNPLRYGALVTKRCARTAVVLVWVVSAAVSFAPIMSQWWRVGADAEAQRCHSNPRCCAFASNMPYVLLSSSVSFYLPLLVMLFVYARVFVVATRQLRLLRGELGRFPPEESPPAPSRSLAPAPVGTCAPPEGVPACGRRPARLLPLREHRALCTLGLIMGTFTLCWLPFFLANVLRALGGPSLVPGPAFLALNWLGYANSAFNPLIYCRSPDFRSAFRRLLCRCGRRLPPEPCAAARPALFPSGVPAARSSPAQPRLCQRLDGASWGVS | |||||||||||||||||||||||||
| 1 | 3sn6R | 0.51 | 0.41 | 0.78 | 2.27 | Download | EAAVNLAKSRWYNQTPNRAKRVITTFRTGTWDAEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAIN----CYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR---------------------------------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNL-IRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLC----------------------------------------------- | |||||||||||||||||||
| 2 | 5uenA | 0.27 | 0.27 | 0.82 | 4.55 | Download | ---------------------------------SAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYFH--TCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLSAVEGSMGEPVIKCEFEKVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDNVKDAGKVKEAQAAAEQLKTTRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCHKPSILTYIAIFLTHGNSAMNPIVYARIQKFRVTFLKIWNDHFRCQPLE-------------------------------------- | |||||||||||||||||||
| 3 | 5uenA | 0.27 | 0.27 | 0.83 | 3.20 | Download | -------------------------------SISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYF--HTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLSAVERAWAAAGSMGEPCEFISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNVKDALTKMRAAALDAPEMKVGQRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCHKPSILTYIAIFLTHGNSAMNPIVYAFRQKFRVTFLKIWNDHFRCQPLEVLF----------------------------------- | |||||||||||||||||||
| 4 | 2rh1 | 0.49 | 0.42 | 0.80 | 1.58 | Download | -------------------------------DEVW-VVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRAT-HQEAINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDEGLRLDTEGYYTTKSPGRTNGVIQKAKSRRTGTWDAYKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDN-LIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLCL---------------------------------------------- | |||||||||||||||||||
| 5 | 3d4s | 0.48 | 0.41 | 0.80 | 1.23 | Download | -----------------------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATH-QEAINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDEGLKDTEGYLDGRNTNGVIKDKPEMLQQKSTGTWDAYKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNL-IRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLCL---------------------------------------------- | |||||||||||||||||||
| 6 | 5uenA | 0.27 | 0.27 | 0.83 | 2.91 | Download | ---------------------------------SAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIGPQTYFH--TCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLSAVERAWAAAGSMGEPVIVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDNVKDAGKVKEAQAAAEQLKTTRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCHKPSILTYIAIFLTHGNSAMNPIVYARIQKFRVTFLKIWNDHFRCQPLEVLF----------------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.22 | 0.22 | 0.74 | 1.73 | Download | -------------------------------VPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYT-LEGVLCNCSFDYISRD-STTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLN---------------------------AKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSSHPKFREAISQTFPW---------------------------------------------- | |||||||||||||||||||
| 8 | 2rh1A | 0.49 | 0.42 | 0.80 | 5.01 | Download | --------------------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRAT-HQEAINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLRMLQQKRWDEAAVNLAKSRWYNQTPNRAKRVITTFRTGTWDAYKFC---LKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQ-DNLIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLCL---------------------------------------------- | |||||||||||||||||||
| 9 | 3sn6R | 0.56 | 0.40 | 0.70 | 2.69 | Download | ---------------------------------EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAI----NCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR---------------------------------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQD-NLIRKEVYILLNWIGYVNSGFNPLIYCRSPDFRIAFQELLC----------------------------------------------- | |||||||||||||||||||
| 10 | 4bvnA | 0.56 | 0.41 | 0.71 | 3.23 | Download | -------------------------------LSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWR-DEDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRKRKTS---------------------------------------RVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRD-LVPKWLFVAFNWLGYANSAMNPIILCRSPDFRKAFKRLLA----------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
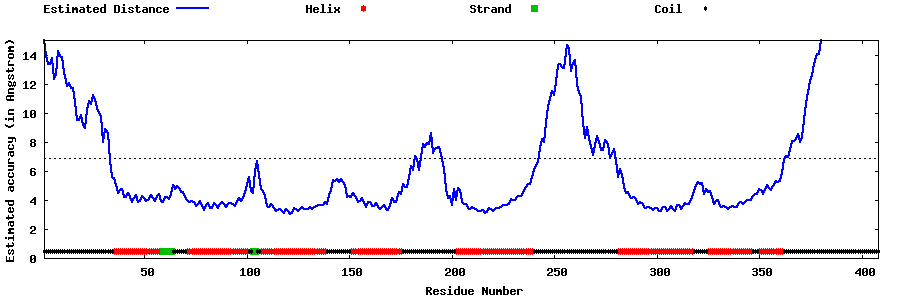
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||