

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MGIPGLEGLHTWISIPFSFMYIVAVAGNIFLIFLIMTERSLHEPMYLFLSMLASADFLLATAAAPKVLAILWFHSMDISFGSCVSQMFFIHFIFVAESAILLAMAFDRYVAICYPLRYTILTSSAVRKIGIAAVVRSFFICCPFIFLVYRLTYCGRNIIPHSYCEHIARLACGNINVNIIYGLTVALLSTGLDIVLIIISYTMILHSVFQISSWAARFKALSTCGSHICVIFMFYTPAFFSFLAHRFGGKTIPHHIHILVGSLYVLVPPMLNPIIYGVKTKQIKDRVILLFSPISVC | |
| CCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCCC | |
| 999995668799999999999999999999999995788753218999999999999899856999999995699867887899999999999999999999998231552046642342488899999999999999999889999973899999944706579999885248368799999999999999899999999999999826888758889854159999999999998999986630468889835899999999738876468112164799999999983277789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MGIPGLEGLHTWISIPFSFMYIVAVAGNIFLIFLIMTERSLHEPMYLFLSMLASADFLLATAAAPKVLAILWFHSMDISFGSCVSQMFFIHFIFVAESAILLAMAFDRYVAICYPLRYTILTSSAVRKIGIAAVVRSFFICCPFIFLVYRLTYCGRNIIPHSYCEHIARLACGNINVNIIYGLTVALLSTGLDIVLIIISYTMILHSVFQISSWAARFKALSTCGSHICVIFMFYTPAFFSFLAHRFGGKTIPHHIHILVGSLYVLVPPMLNPIIYGVKTKQIKDRVILLFSPISVC | |
| 741022132020001102201220332121000002115100100020001002001000201002000000020440304000000120031033011001100300000002101000003410010000003213012202101021032045200000001000100033030011100200331333323103201120020002002460132013001001000100323133101000104330021000100331013013300300002034004100300254638 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCCC MGIPGLEGLHTWISIPFSFMYIVAVAGNIFLIFLIMTERSLHEPMYLFLSMLASADFLLATAAAPKVLAILWFHSMDISFGSCVSQMFFIHFIFVAESAILLAMAFDRYVAICYPLRYTILTSSAVRKIGIAAVVRSFFICCPFIFLVYRLTYCGRNIIPHSYCEHIARLACGNINVNIIYGLTVALLSTGLDIVLIIISYTMILHSVFQISSWAARFKALSTCGSHICVIFMFYTPAFFSFLAHRFGGKTIPHHIHILVGSLYVLVPPMLNPIIYGVKTKQIKDRVILLFSPISVC | |||||||||||||||||||||||||
| 1 | 3emlA | 0.17 | 0.25 | 0.93 | 2.64 | Download | -----IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACL---------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.23 | 0.91 | 2.13 | Download | -------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSV--------------CSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------ | |||||||||||||||||||
| 3 | 3pblA | 0.20 | 0.21 | 0.87 | 1.85 | Download | ------------YALSYCALILAIVFGNGLVCMAVLKERALQTTTNYLVVSLAVADLLVATLVMPWVVYLEVTG-GVWNFSCCDVFVTLDVMMCTASIWNLCAISIDRYTAVVMPVHYQHGSCRRVALMITAVWVLAFAVSCPLLF--GFNTTCSIS--------------------NPDFVIYSSVVSFYLPFGVTVLVYARIYVVLKQRRVPLREKKATQMVAIVLGAFIVCWLPFFLTHVLNTHCQTCHVSPELYSATTWLGYVNSALNPVIYTTFNIEFRKAFLKILSC---- | |||||||||||||||||||
| 4 | 3uon | 0.19 | 0.25 | 0.93 | 1.56 | Download | ----TF--EVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQ-------FFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPN-TVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----- | |||||||||||||||||||
| 5 | 2rh1 | 0.20 | 0.24 | 0.94 | 1.12 | Download | -----DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYSLLTKNKARVIILMVWIVSGLTSFLPIQM-H---WYRATHQEAINCYA--EETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNILKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVY-ILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL---- | |||||||||||||||||||
| 6 | 3emlA | 0.17 | 0.25 | 0.92 | 2.83 | Download | --------GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV---------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.25 | 0.88 | 1.69 | Download | -------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSARTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVNT------------------DHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH----L-AI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 8 | 4buoA | 0.15 | 0.20 | 0.95 | 3.18 | Download | LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYI---------SDEQWTYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL------ | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.25 | 0.91 | 3.07 | Download | -----I--GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISF----CAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVV----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFF-----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 10 | 4gpoA | 0.19 | 0.22 | 0.93 | 4.88 | Download | -----LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP-DWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
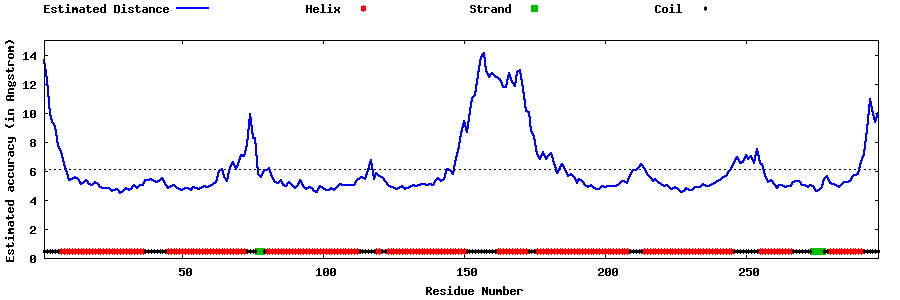
| Generated 3D models | Estimated local accuracy of models | ||
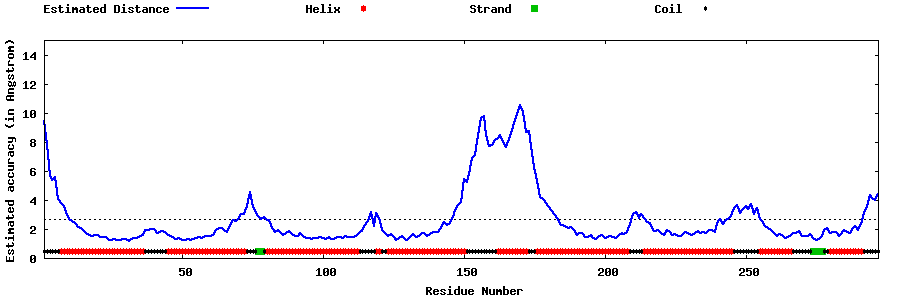 | |||
|
|
|||
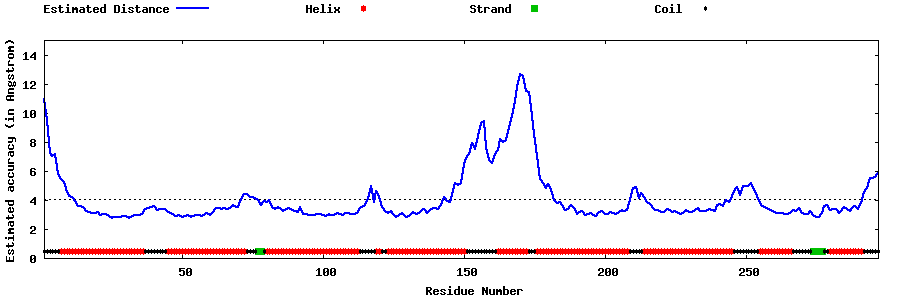 | |||
|
| |||
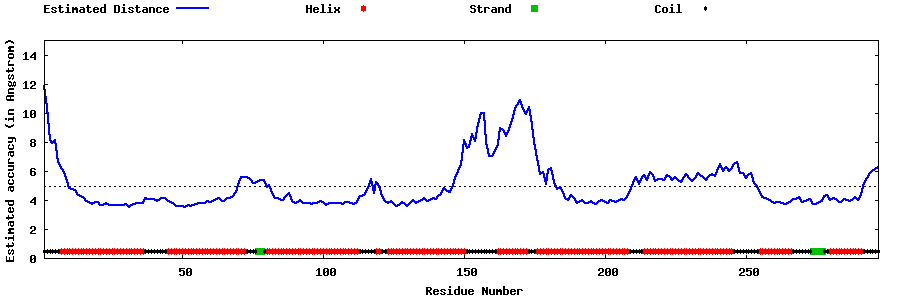 | |||
|
| |||
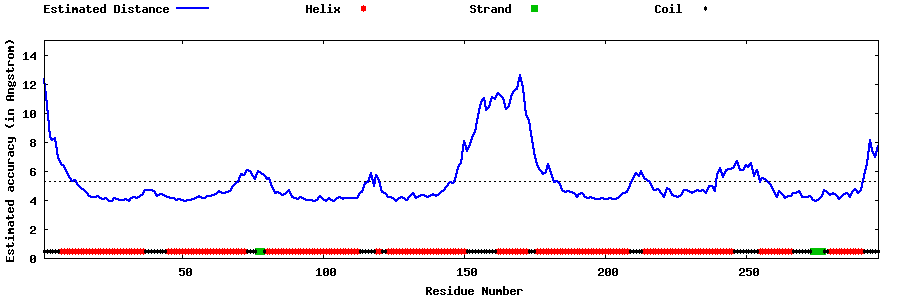 | |||
|
| |||
 | |||
|
| |||