

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MNATGTPVAPESCQQLAAGGHSRLIVLHYNHSGRLAGRGGPEDGGLGALRGLSVAASCLVVLENLLVLAAITSHMRSRRWVYYCLVNITLSDLLTGAAYLANVLLSGARTFRLAPAQWFLREGLLFTALAASTFSLLFTAGERFATMVRPVAESGATKTSRVYGFIGLCWLLAALLGMLPLLGWNCLCAFDRCSSLLPLYSKRYILFCLVIFAGVLATIMGLYGAIFRLVQASGQKAPRPAARRKARRLLKTVLMILLAFLVCWGPLFGLLLADVFGSNLWAQEYLRGMDWILALAVLNSAVNPIIYSFRSREVCRAVLSFLCCGCLRLGMRGPGDCLARAVEAHSGASTTDSSLRPRDSFRGSRSLSFRMREPLSSISSVRSI | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCCCCSSSCCCCCCSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 999999999765464322578874204467778878778997415999999999999998997686414421379889489999999999999999999999999960454447434332889999999999999999999708886046878755329999999999999999999999986050107987425344465404675448999999999999999999999987541463123888889989999999999999999999999996887677627999999999999999884999996499999999999477888877788987656556677788888887788877778888877778999986667879 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MNATGTPVAPESCQQLAAGGHSRLIVLHYNHSGRLAGRGGPEDGGLGALRGLSVAASCLVVLENLLVLAAITSHMRSRRWVYYCLVNITLSDLLTGAAYLANVLLSGARTFRLAPAQWFLREGLLFTALAASTFSLLFTAGERFATMVRPVAESGATKTSRVYGFIGLCWLLAALLGMLPLLGWNCLCAFDRCSSLLPLYSKRYILFCLVIFAGVLATIMGLYGAIFRLVQASGQKAPRPAARRKARRLLKTVLMILLAFLVCWGPLFGLLLADVFGSNLWAQEYLRGMDWILALAVLNSAVNPIIYSFRSREVCRAVLSFLCCGCLRLGMRGPGDCLARAVEAHSGASTTDSSLRPRDSFRGSRSLSFRMREPLSSISSVRSI | |
| 763443333253143234332332021210202424445324411100000100100210230130000000103300100000000001001101312110010003211200211010100011000100120000002000000010032332233100000000001101310310000001213342000011132100000002313321220020011001101310443644643331230010000001000000211200000000033230310010010010001100010000000105400300110000120325354454444444444444343544243464444446243445533444444647 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCCCCSSSCCCCCCSSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MNATGTPVAPESCQQLAAGGHSRLIVLHYNHSGRLAGRGGPEDGGLGALRGLSVAASCLVVLENLLVLAAITSHMRSRRWVYYCLVNITLSDLLTGAAYLANVLLSGARTFRLAPAQWFLREGLLFTALAASTFSLLFTAGERFATMVRPVAESGATKTSRVYGFIGLCWLLAALLGMLPLLGWNCLCAFDRCSSLLPLYSKRYILFCLVIFAGVLATIMGLYGAIFRLVQASGQKAPRPAARRKARRLLKTVLMILLAFLVCWGPLFGLLLADVFGSNLWAQEYLRGMDWILALAVLNSAVNPIIYSFRSREVCRAVLSFLCCGCLRLGMRGPGDCLARAVEAHSGASTTDSSLRPRDSFRGSRSLSFRMREPLSSISSVRSI | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.27 | 0.22 | 0.75 | 2.79 | Download | -----------------------------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSF--SDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGK--MNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------------------------------------------------------------- | |||||||||||||||||||
| 2 | 4z34A | 0.33 | 0.28 | 0.79 | 3.70 | Download | --------------NEPQCFYNESIAFFYNRSGKHLATEWNT--VSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQ-LHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWETLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDVLAYEKFF---LLLAEFNSAMNPIIYSYRDKEMSATFRQIL-------------------------------------------------------------- | |||||||||||||||||||
| 3 | 5tgzA | 0.26 | 0.24 | 0.76 | 3.20 | Download | -------------------------GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFP--HIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA---PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGK--MNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------------------------------------------------------------- | |||||||||||||||||||
| 4 | 4z34 | 0.34 | 0.28 | 0.79 | 1.57 | Download | --------------NEPQCFYNESIAFFYNRSGKHLATEWNT--VSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFR-MQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLERKAMYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQC-D-V-LAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG------------------------------------------------------------- | |||||||||||||||||||
| 5 | 4ib4 | 0.17 | 0.17 | 0.75 | 1.18 | Download | ---------------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------------------------------------------------- | |||||||||||||||||||
| 6 | 5tjvA | 0.27 | 0.22 | 0.75 | 3.01 | Download | -----------------------------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAG--KRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGK--MNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------------------------------------------------------------- | |||||||||||||||||||
| 7 | 4z34 | 0.34 | 0.28 | 0.78 | 1.79 | Download | -------------------FYNESIAFFYNRSGKHLATEW--NTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFR-MQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLERKAMYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDV---LAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG------------------------------------------------------------- | |||||||||||||||||||
| 8 | 1u19A | 0.20 | 0.22 | 0.86 | 4.58 | Download | MNGTEGP---------------NFYVPFSNKTGVVRSPFEAPPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFG-ENHAIMGVAFTWVMALACAAPPLVGWSRYGMQ--CSCGIDYYTPHEETMFVVHFIIPLIVIFFCYGQLVFTVKEAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFG--PIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA----------------------------------- | |||||||||||||||||||
| 9 | 5tjvA | 0.27 | 0.22 | 0.75 | 2.95 | Download | -----------------------------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSF--SDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKM--NKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------------------------------------------------------------- | |||||||||||||||||||
| 10 | 5g53A | 0.23 | 0.21 | 0.73 | 2.92 | Download | ---------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS-TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCCGEGQCLFEDVVPMNYMVYFNFFCVLVPLLLMLGVYLRIFLAARRQLKQM----TLQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAP-LWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLENLY----------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
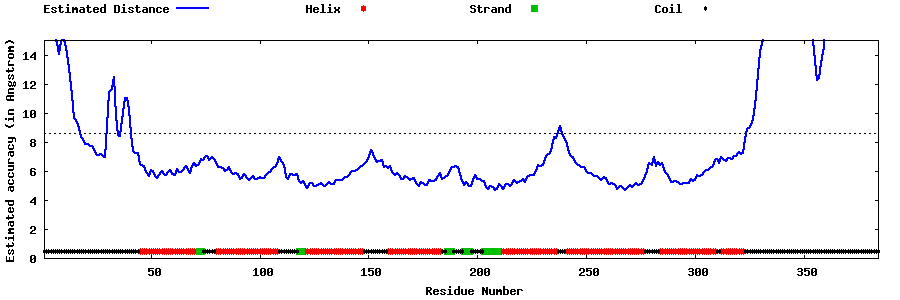
| Generated 3D models | Estimated local accuracy of models | ||
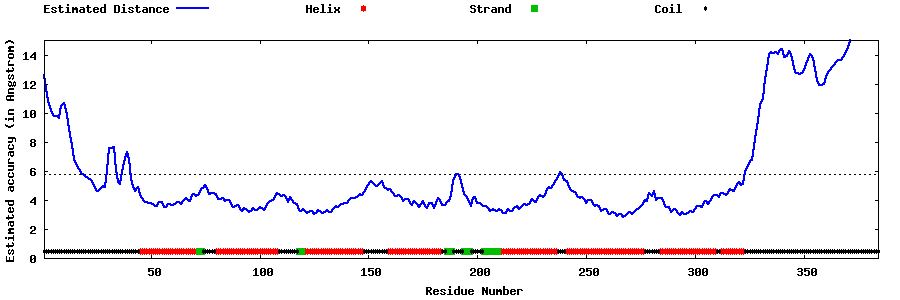 | |||
|
|
|||
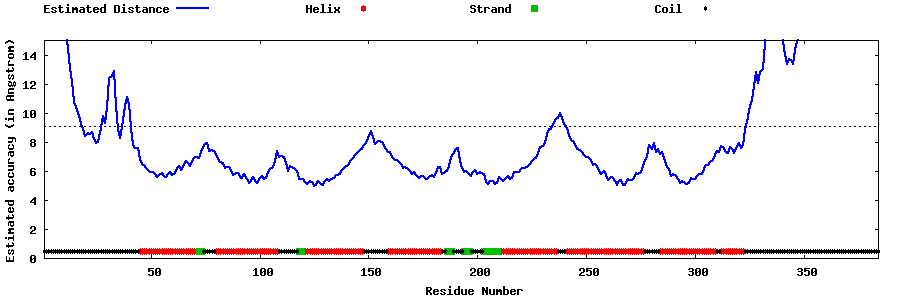 | |||
|
| |||
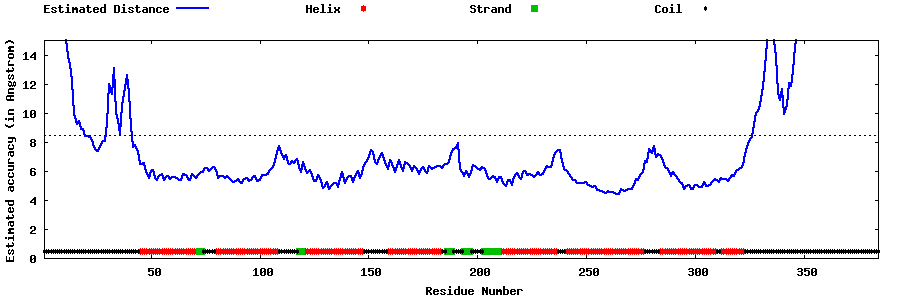 | |||
|
| |||
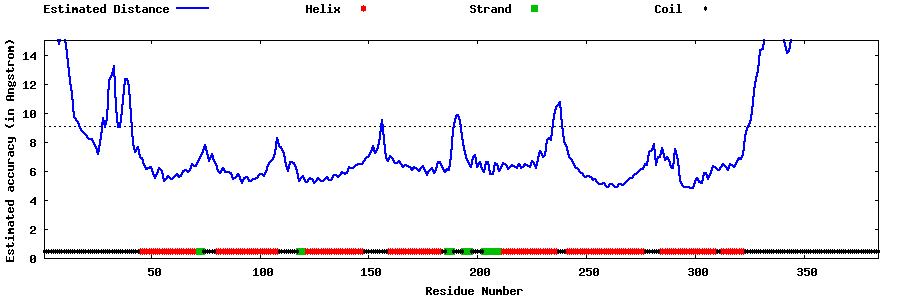 | |||
|
| |||
 | |||
|
| |||