

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MADAQNISLDSPGSVGAVAVPVVFALIFLLGTVGNGLVLAVLLQPGPSAWQEPGSTTDLFILNLAVADLCFILCCVPFQATIYTLDAWLFGALVCKAVHLLIYLTMYASSFTLAAVSVDRYLAVRHPLRSRALRTPRNARAAVGLVWLLAALFSAPYLSYYGTVRYGALELCVPAWEDARRRALDVATFAAGYLLPVAVVSLAYGRTLRFLWAAVGPAGAAAAEARRRATGRAGRAMLAVAALYALCWGPHHALILCFWYGRFAFSPATYACRLASHCLAYANSCLNPLVYALASRHFRARFRRLWPCGRRRRHRARRALRRVRPASSGPPGCPGDARPSGRLLAGGGQGPEPREGPVHGGEAARGPE | |
| CCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99987788887278999999999999999998877831778886376535677879999999999999999999889999999669988971788789999999999999999999998588887007311053477788763799999999997999981489969988982779858999999999999999999999999999999995578987620478877714568799999999999998999999999998434441899999999999999999499999998099999999999482467887766765677888899998999877765010799988887778887786789999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MADAQNISLDSPGSVGAVAVPVVFALIFLLGTVGNGLVLAVLLQPGPSAWQEPGSTTDLFILNLAVADLCFILCCVPFQATIYTLDAWLFGALVCKAVHLLIYLTMYASSFTLAAVSVDRYLAVRHPLRSRALRTPRNARAAVGLVWLLAALFSAPYLSYYGTVRYGALELCVPAWEDARRRALDVATFAAGYLLPVAVVSLAYGRTLRFLWAAVGPAGAAAAEARRRATGRAGRAMLAVAALYALCWGPHHALILCFWYGRFAFSPATYACRLASHCLAYANSCLNPLVYALASRHFRARFRRLWPCGRRRRHRARRALRRVRPASSGPPGCPGDARPSGRLLAGGGQGPEPREGPVHGGEAARGPE | |
| 44244423263222000000021112001102311200000000024432330100000001000300100000010000000014310002000100000131000000000000010001000000202332123100000000111010001000000202537412000020353121000001033112310100020001000101313344644434443342200000000000000000222000000100141333200000100000000000003000100014300400220020024344445443444444444454353544344433445444453464434446546648 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCCCCCCCCCCCCCCCCCCCCC MADAQNISLDSPGSVGAVAVPVVFALIFLLGTVGNGLVLAVLLQPGPSAWQEPGSTTDLFILNLAVADLCFILCCVPFQATIYTLDAWLFGALVCKAVHLLIYLTMYASSFTLAAVSVDRYLAVRHPLRSRALRTPRNARAAVGLVWLLAALFSAPYLSYYGTVRYGALELCVPAWEDARRRALDVATFAAGYLLPVAVVSLAYGRTLRFLWAAVGPAGAAAAEARRRATGRAGRAMLAVAALYALCWGPHHALILCFWYGRFAFSPATYACRLASHCLAYANSCLNPLVYALASRHFRARFRRLWPCGRRRRHRARRALRRVRPASSGPPGCPGDARPSGRLLAGGGQGPEPREGPVHGGEAARGPE | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.27 | 0.26 | 0.81 | 3.32 | Download | ----SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVR-----YTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.24 | 0.22 | 0.80 | 3.76 | Download | -----PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILIL-----INYKRLKSMTDIYLLNLAISDLFFLLT-VPFWAHYAA-AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMGVGKDQFEEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.28 | 0.26 | 0.81 | 3.64 | Download | ----SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY-----TKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.23 | 0.24 | 0.80 | 1.56 | Download | ----------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLE-----KKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIK-GIETNPNNITCVLTKER--FGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.30 | 0.29 | 0.77 | 1.17 | Download | ------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY-----TKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNTPYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA-----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------------------------ | |||||||||||||||||||
| 6 | 4n6hA | 0.28 | 0.26 | 0.81 | 3.50 | Download | ----SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVR-----YTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG---SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.23 | 0.24 | 0.76 | 1.72 | Download | ----------------VVFIVLVAGSLSLVTIIGNILVMVSIKVN-----RHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVTVEDGECYIQFFSN--AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGADAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCI---PNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.29 | 0.26 | 0.80 | 5.01 | Download | ---------GSPSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY-----TKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.27 | 0.26 | 0.81 | 3.37 | Download | ----SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVR-----YTKMKTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.28 | 0.25 | 0.79 | 4.49 | Download | -GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVR-----YTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS---KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
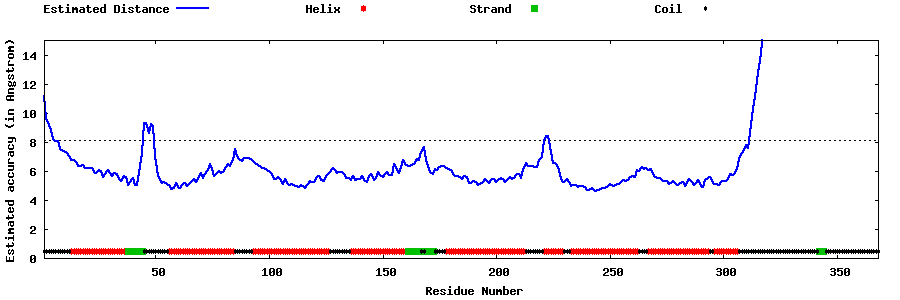
| Generated 3D models | Estimated local accuracy of models | ||
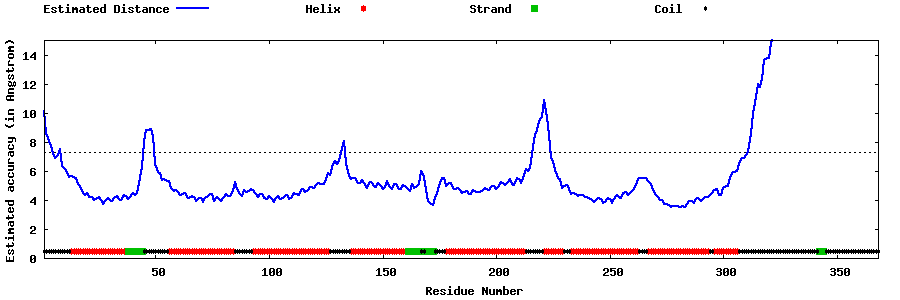 | |||
|
|
|||
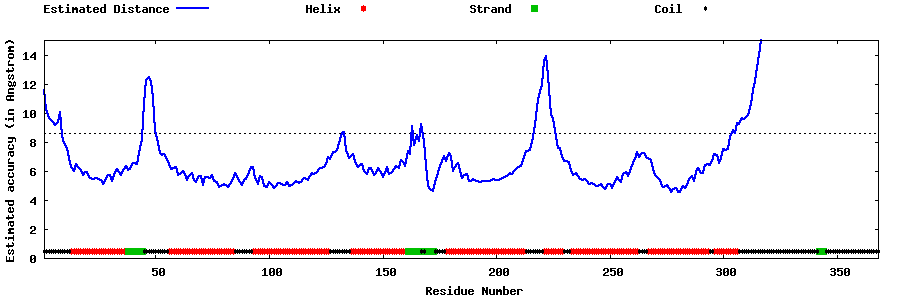 | |||
|
| |||
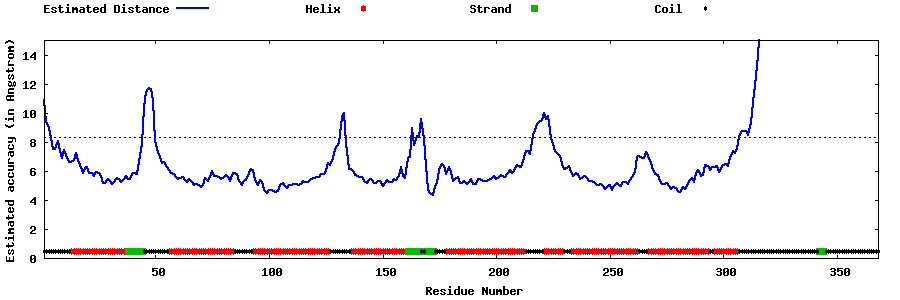 | |||
|
| |||
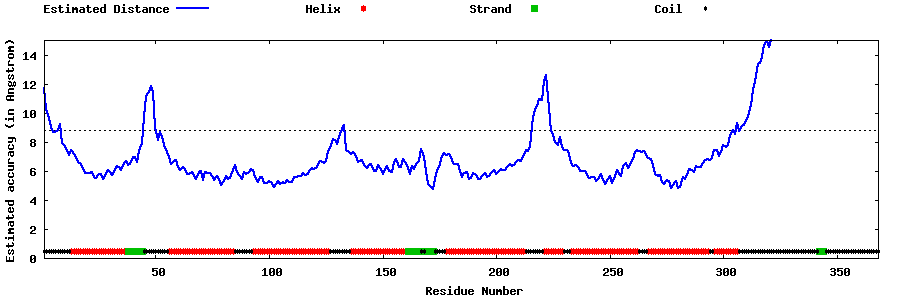 | |||
|
| |||
 | |||
|
| |||