

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MWQEYYFLNVFFPLLKVCCLTINSHVVILLPWECYHLIWKILPYIGTTVGSMEEYNTSSTDFTFMGLFNRKETSGLIFAIISIIFFTALMANGVMIFLIQTDLRLHTPMYFLLSHLSLIDMMYISTIVPKMLVNYLLDQRTISFVGCTAQHFLYLTLVGAEFFLLGLMAYDRYVAICNPLRYPVLMSRRVCWMIIAGSWFGGSLDGFLLTPITMSFPFCNSREINHFFCEAPAVLKLACADTALYETVMYVCCVLMLLIPFSVVLASYARILTTVQCMSSVEGRKKAFATCSSHMTVVSLFYGAAMYTYMLPHSYHKPAQDKVLSVFYTILTPMLNPLIYSLRNKDVTGALKRALGRFKGPQRVSGGVF | |
| CCCCCHHHHHHHHHHHHHHSSCCSSSSSSSSCCHHHHHHHHCCCHHHHHHHHHCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSSHHHHHHHHCCHHHCCCCHHHHHHHHHHHHHCCCHHHCCCCCC | |
| 974204345327888786041321489998600122677633203458888743886313669965899944488999999999999999889997989757775573889987899998753600589999985369968848999999999999999999999999765056305454588415886999999999999999999999999135889989678761482888888713751989999999999999989999999999999981387703253076607878997999987577515288999997669689821740776320254214649999999999985346232046889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MWQEYYFLNVFFPLLKVCCLTINSHVVILLPWECYHLIWKILPYIGTTVGSMEEYNTSSTDFTFMGLFNRKETSGLIFAIISIIFFTALMANGVMIFLIQTDLRLHTPMYFLLSHLSLIDMMYISTIVPKMLVNYLLDQRTISFVGCTAQHFLYLTLVGAEFFLLGLMAYDRYVAICNPLRYPVLMSRRVCWMIIAGSWFGGSLDGFLLTPITMSFPFCNSREINHFFCEAPAVLKLACADTALYETVMYVCCVLMLLIPFSVVLASYARILTTVQCMSSVEGRKKAFATCSSHMTVVSLFYGAAMYTYMLPHSYHKPAQDKVLSVFYTILTPMLNPLIYSLRNKDVTGALKRALGRFKGPQRVSGGVF | |
| 613421111000110100003030300000123322101200332433143046210120000000003325010000120333233133323200100100130000001102000000011000200200000016532000000220010000001101200020020000000200100020033000000110020003003100010000100332301000001100020001002200210123113313311210220112000000103145123101000000000000122000000010314334633100001102310330030000104202300220023232344035324 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCHHHHHHHHHHHHHHSSCCSSSSSSSSCCHHHHHHHHCCCHHHHHHHHHCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSSHHHHHHHHCCHHHCCCCHHHHHHHHHHHHHCCCHHHCCCCCC MWQEYYFLNVFFPLLKVCCLTINSHVVILLPWECYHLIWKILPYIGTTVGSMEEYNTSSTDFTFMGLFNRKETSGLIFAIISIIFFTALMANGVMIFLIQTDLRLHTPMYFLLSHLSLIDMMYISTIVPKMLVNYLLDQRTISFVGCTAQHFLYLTLVGAEFFLLGLMAYDRYVAICNPLRYPVLMSRRVCWMIIAGSWFGGSLDGFLLTPITMSFPFCNSREINHFFCEAPAVLKLACADTALYETVMYVCCVLMLLIPFSVVLASYARILTTVQCMSSVEGRKKAFATCSSHMTVVSLFYGAAMYTYMLPHSYHKPAQDKVLSVFYTILTPMLNPLIYSLRNKDVTGALKRALGRFKGPQRVSGGVF | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.19 | 0.75 | 2.98 | Download | ----------------------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAA-KSLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------- | |||||||||||||||||||
| 2 | 5tgzA | 0.16 | 0.18 | 0.76 | 2.04 | Download | -----------------------------------------------------ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEK--------LQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM--------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.17 | 0.17 | 0.71 | 1.89 | Download | --------------------------------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH-----------------ILYTVYSTVGAFY---FPTLLLIALYGRIYVEARSQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----------- | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.17 | 0.76 | 1.53 | Download | ---------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------ | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.23 | 0.88 | 1.21 | Download | PDSPEMK--DFRHGFDILVGQID-------DALKL-ANEGKVKEQAAAEQLKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-------------- | |||||||||||||||||||
| 6 | 3emlA | 0.19 | 0.19 | 0.75 | 3.20 | Download | -----------------------------------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------- | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.17 | 0.71 | 1.71 | Download | -----------------------------------------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LA-IFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----------- | |||||||||||||||||||
| 8 | 4buoA | 0.19 | 0.18 | 0.77 | 2.92 | Download | -------------------------------------------------------NSDL------DVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGTPI----------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTNALVYVSAAINPILYNLVSANFRQVFLSTL-------------- | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.19 | 0.76 | 4.05 | Download | ----------------------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQSQGCGEGQVAC--------LFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------- | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.18 | 0.80 | 4.55 | Download | -------------------------------------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLPFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPFKYF | |||||||||||||||||||
| ||||||||||||||||||||||||||
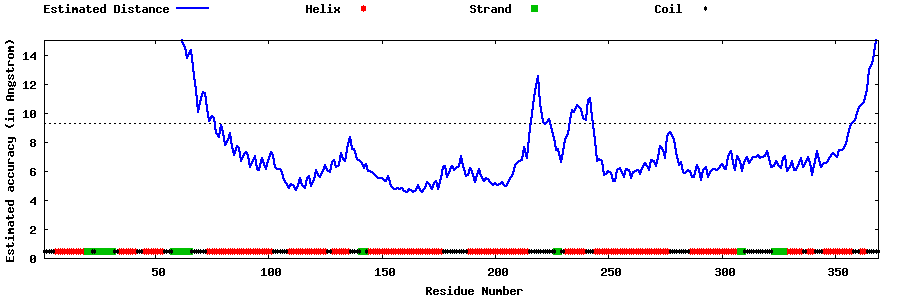
| Generated 3D models | Estimated local accuracy of models | ||
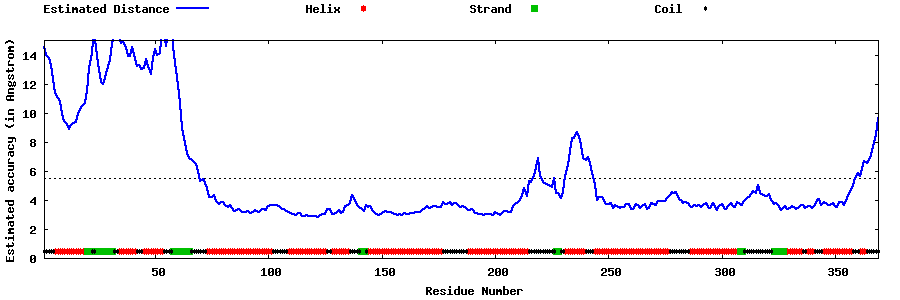 | |||
|
|
|||
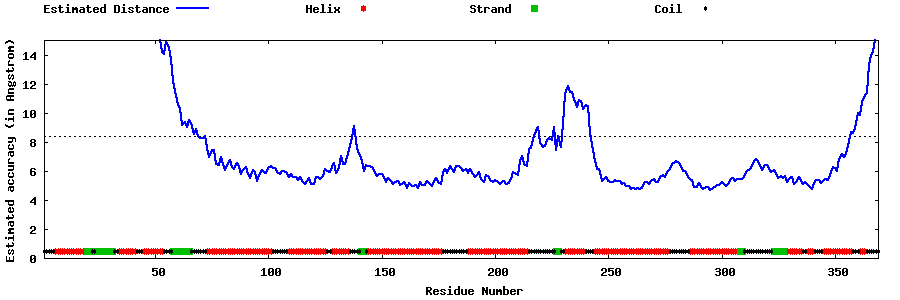 | |||
|
| |||
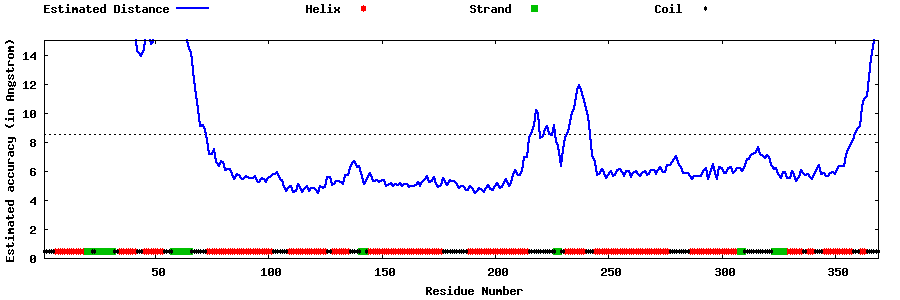 | |||
|
| |||
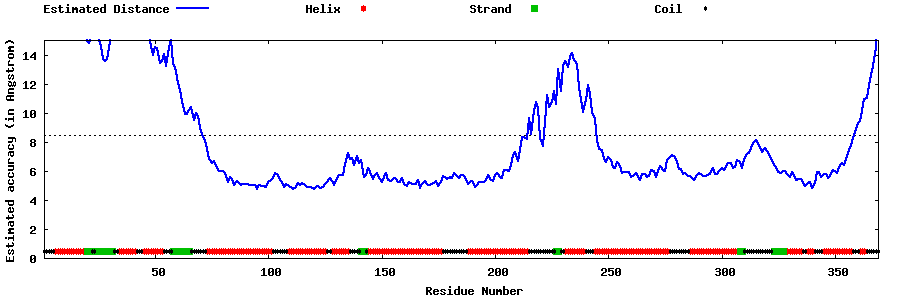 | |||
|
| |||
 | |||
|
| |||