

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MNVSGCPGAGNASQAGGGGGWHPEAVIVPLLFALIFLVGTVGNTLVLAVLLRGGQAVSTTNLFILNLGVADLCFILCCVPFQATIYTLDGWVFGSLLCKAVHFLIFLTMHASSFTLAAVSLDRYLAIRYPLHSRELRTPRNALAAIGLIWGLSLLFSGPYLSYYRQSQLANLTVCHPAWSAPRRRAMDICTFVFSYLLPVLVLGLTYARTLRYLWRAVDPVAAGSGARRAKRKVTRMILIVAALFCLCWMPHHALILCVWFGQFPLTRATYALRILSHLVSYANSCVNPIVYALVSKHFRKGFRTICAGLLGRAPGRASGRVCAAARGTHSGSVLERESSDLLHMSEAAGALRPCPGASQPCILEPCPGPSWQGPKAGDSILTVDVA | |
| CCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 988999999888877776653769998999999999999866885202013178888699999999999999999998899999997289779628888899999999999999999999983776413175110104676877638999999999979988746898489779808898349999999999999999999999999999999946689975115666730249999999999999999999999999997103330799999999999999999599999998199999999999765437887666777566667777787765666656564666655578999998757899999745688775765444469 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MNVSGCPGAGNASQAGGGGGWHPEAVIVPLLFALIFLVGTVGNTLVLAVLLRGGQAVSTTNLFILNLGVADLCFILCCVPFQATIYTLDGWVFGSLLCKAVHFLIFLTMHASSFTLAAVSLDRYLAIRYPLHSRELRTPRNALAAIGLIWGLSLLFSGPYLSYYRQSQLANLTVCHPAWSAPRRRAMDICTFVFSYLLPVLVLGLTYARTLRYLWRAVDPVAAGSGARRAKRKVTRMILIVAALFCLCWMPHHALILCVWFGQFPLTRATYALRILSHLVSYANSCVNPIVYALVSKHFRKGFRTICAGLLGRAPGRASGRVCAAARGTHSGSVLERESSDLLHMSEAAGALRPCPGASQPCILEPCPGPSWQGPKAGDSILTVDVA | |
| 303032323323133344332200000002110200010131120000000003402100010010003001000100000000100133000010001000001110000000000000100010000002023311231000000001100100010000002026354110000201541210000010231113101000200000011013234336455533431100000000000000000212000000100231331200000100000000100000000000004301400240021024344544444335444434444434344443343454544444344544433444344333634534743333628 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MNVSGCPGAGNASQAGGGGGWHPEAVIVPLLFALIFLVGTVGNTLVLAVLLRGGQAVSTTNLFILNLGVADLCFILCCVPFQATIYTLDGWVFGSLLCKAVHFLIFLTMHASSFTLAAVSLDRYLAIRYPLHSRELRTPRNALAAIGLIWGLSLLFSGPYLSYYRQSQLANLTVCHPAWSAPRRRAMDICTFVFSYLLPVLVLGLTYARTLRYLWRAVDPVAAGSGARRAKRKVTRMILIVAALFCLCWMPHHALILCVWFGQFPLTRATYALRILSHLVSYANSCVNPIVYALVSKHFRKGFRTICAGLLGRAPGRASGRVCAAARGTHSGSVLERESSDLLHMSEAAGALRPCPGASQPCILEPCPGPSWQGPKAGDSILTVDVA | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.28 | 0.26 | 0.80 | 2.82 | Download | LKTTRYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------- | |||||||||||||||||||
| 2 | 2ziy | 0.19 | 0.20 | 0.90 | 1.49 | Download | -DLRDNETWWYNPSIEFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLGVLCNCSFDYISRDTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLE--WVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETED-----DKD--------AET--EIPAGESSDA--------------APSADAAQMKE-------- | |||||||||||||||||||
| 3 | 4n6hA | 0.29 | 0.26 | 0.76 | 3.85 | Download | -----------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATS-TLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG-SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK------------------------------------------------------------------------------ | |||||||||||||||||||
| 4 | 4zwjA | 0.17 | 0.28 | 0.99 | 2.69 | Download | SNATG-VVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRF-GENHAIMGVAFTWVMALACAAPPLAWSRYIPEGLQCSCGIDYYTLKNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAAAQQQESATTQKAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQGSC--FGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICCGKNVIFKKVSRDKGKRDYVDHVSQVEPVDGVVLVDPELVKGKKVYVTLTCAFRYGQEDIDVMGLTFRRDLYFSRVQVY | |||||||||||||||||||
| 5 | 4n6hA | 0.29 | 0.26 | 0.77 | 3.64 | Download | ------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLS-GSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4ib4 | 0.23 | 0.23 | 0.75 | 1.49 | Download | ------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIK-GIETNPNNITCVLTKER--FGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------------------------------------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.31 | 0.26 | 0.73 | 1.17 | Download | --------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA-----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------------------------------------------ | |||||||||||||||||||
| 8 | 4n6hA | 0.29 | 0.26 | 0.76 | 3.34 | Download | --------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL-SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 3uon | 0.23 | 0.22 | 0.72 | 1.67 | Download | ------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVTVEDGECYIQFFSN--AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC---IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------------------------------------------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.29 | 0.26 | 0.77 | 4.94 | Download | -----------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV-RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
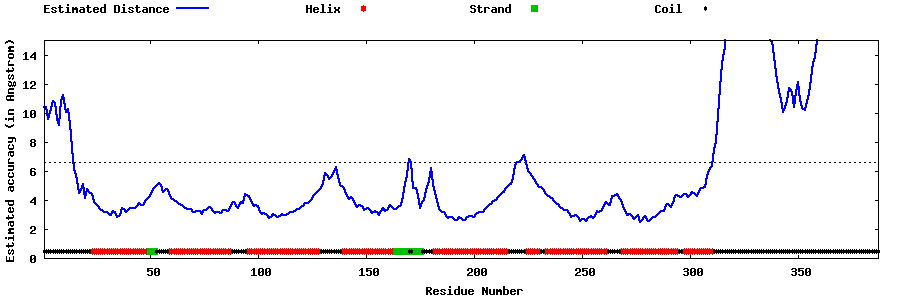 | |||
|
|
|||
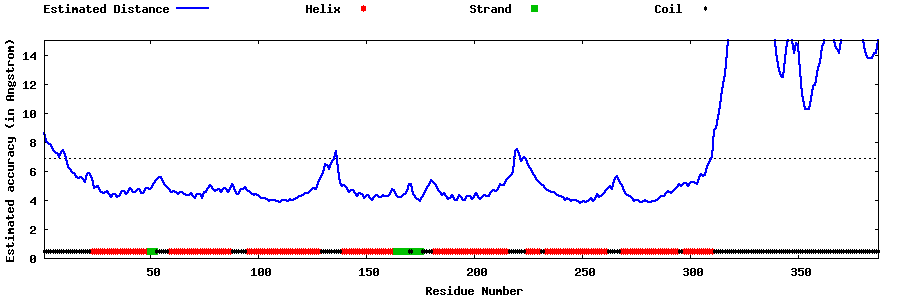 | |||
|
| |||
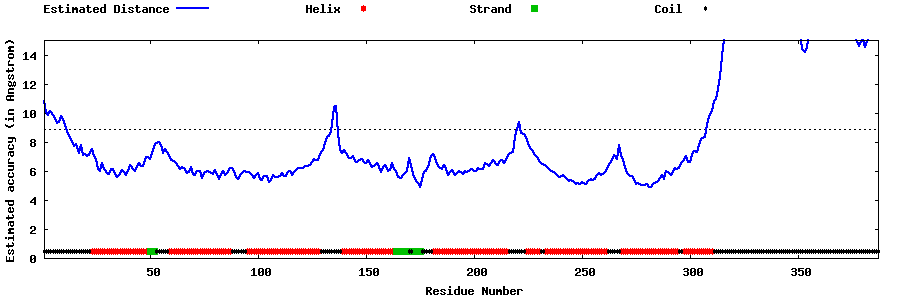 | |||
|
| |||
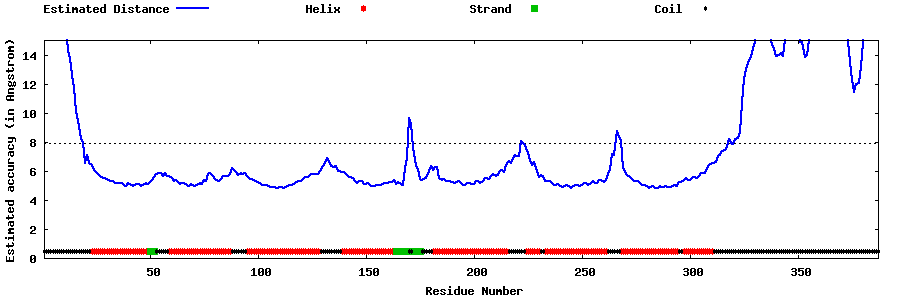 | |||
|
| |||
 | |||
|
| |||