

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MGSPWNGSDGPEGAREPPWPALPPCDERRCSPFPLGALVPVTAVCLCLFVVGVSGNVVTVMLIGRYRDMRTTTNLYLGSMAVSDLLILLGLPFDLYRLWRSRPWVFGPLLCRLSLYVGEGCTYATLLHMTALSVERYLAICRPLRARVLVTRRRVRALIAVLWAVALLSAGPFLFLVGVEQDPGISVVPGLNGTARIASSPLASSPPLWLSRAPPPSPPSGPETAEAAALFSRECRPSPAQLGALRVMLWVTTAYFFLPFLCLSILYGLIGRELWSSRRPLRGPAASGRERGHRQTVRVLLVVVLAFIICWLPFHVGRIIYINTEDSRMMYFSQYFNIVALQLFYLSASINPILYNLISKKYRAAAFKLLLARKSRPRGFHRSRDTAGEVAGDTGGDTVGYTETSANVKTMG | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCCCCCCCCCCSSCCHHHSSSCCCCCCCCCHHHHHHHSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCC | |
| 9899888999777889998888986555677774268999999999999998642365178888606766324899999999999999953999999996798558528898899999999999999999999871500102465331457778776679999999999868977102363267642235676422233321000000012012211121002233200220024642878756777777888887635999999889999998403576556324666666755533335678999998579999999998657753115999999999999999998999999928999999999995656789976566767678868899988558857878700689 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MGSPWNGSDGPEGAREPPWPALPPCDERRCSPFPLGALVPVTAVCLCLFVVGVSGNVVTVMLIGRYRDMRTTTNLYLGSMAVSDLLILLGLPFDLYRLWRSRPWVFGPLLCRLSLYVGEGCTYATLLHMTALSVERYLAICRPLRARVLVTRRRVRALIAVLWAVALLSAGPFLFLVGVEQDPGISVVPGLNGTARIASSPLASSPPLWLSRAPPPSPPSGPETAEAAALFSRECRPSPAQLGALRVMLWVTTAYFFLPFLCLSILYGLIGRELWSSRRPLRGPAASGRERGHRQTVRVLLVVVLAFIICWLPFHVGRIIYINTEDSRMMYFSQYFNIVALQLFYLSASINPILYNLISKKYRAAAFKLLLARKSRPRGFHRSRDTAGEVAGDTGGDTVGYTETSANVKTMG | |
| 7743223243342333132332331445423313000100102101100210130120000000113402100000000001000000000121000002432001010001000000100000000000000000010001002023311231000000000010000001000002033343343232333323333233221221112212223232222112000000222223333322200101110111020000001010200220243444364543443333413001020013400012001000000000003304222000000000003133201211231000134114002404202323444343434434434443333233334332345338 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCCCCCCCCCCSSCCHHHSSSCCCCCCCCCHHHHHHHSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCC MGSPWNGSDGPEGAREPPWPALPPCDERRCSPFPLGALVPVTAVCLCLFVVGVSGNVVTVMLIGRYRDMRTTTNLYLGSMAVSDLLILLGLPFDLYRLWRSRPWVFGPLLCRLSLYVGEGCTYATLLHMTALSVERYLAICRPLRARVLVTRRRVRALIAVLWAVALLSAGPFLFLVGVEQDPGISVVPGLNGTARIASSPLASSPPLWLSRAPPPSPPSGPETAEAAALFSRECRPSPAQLGALRVMLWVTTAYFFLPFLCLSILYGLIGRELWSSRRPLRGPAASGRERGHRQTVRVLLVVVLAFIICWLPFHVGRIIYINTEDSRMMYFSQYFNIVALQLFYLSASINPILYNLISKKYRAAAFKLLLARKSRPRGFHRSRDTAGEVAGDTGGDTVGYTETSANVKTMG | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.27 | 0.22 | 0.73 | 2.32 | Download | -------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQ--------------------------------------------FPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.23 | 0.21 | 0.76 | 4.06 | Download | ----------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHVIAIPINVYKLLAED-WPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDYKGS-------------------------------------YLRICLLHPVQKTAFMQFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQELLSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC---------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.27 | 0.22 | 0.73 | 3.37 | Download | -------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-----------------------AVVCMLQFPSPSWYWDTVTKICVFLFA---------------------FVVPILIITVCYGLMLLRLRSVRLLSG---SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.23 | 0.20 | 0.71 | 1.55 | Download | -------------------------EEQGN------KLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKG-IETNPN-----------------------------------------------NITCVLTKER--FGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN-QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.26 | 0.23 | 0.70 | 1.17 | Download | ---------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVVIE-------------------------------------------CSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.27 | 0.22 | 0.73 | 2.85 | Download | ---------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQ--------------------------------------------FPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG---SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.22 | 0.23 | 0.79 | 1.67 | Download | -------------------------------------PVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDSWVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIF---EMLRIDEGLRLKYKDTEGDKAIGRGVITKVVAGFLQQKRTTFRTGTWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF----------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.28 | 0.22 | 0.72 | 4.21 | Download | -----------------------------GSPGSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG--------------------------------------------AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.27 | 0.22 | 0.73 | 2.30 | Download | -------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG--------------------------------------------AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS---KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.24 | 0.21 | 0.72 | 3.06 | Download | ----------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQG--------------------------------------------SIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRML---SGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPE-TTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
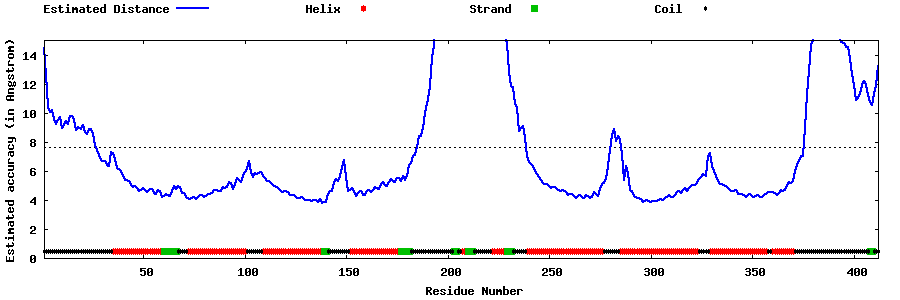 | |||
|
| |||
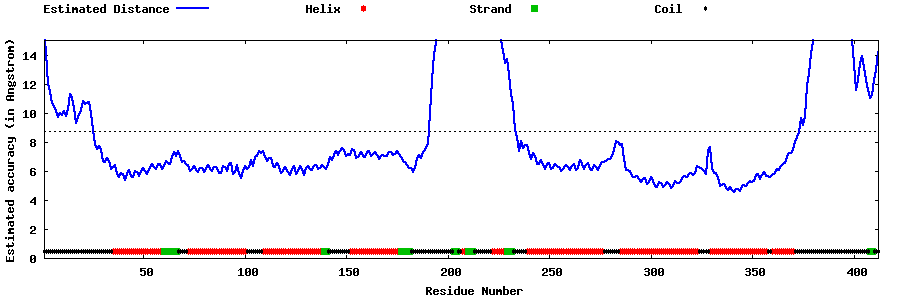 | |||
|
| |||
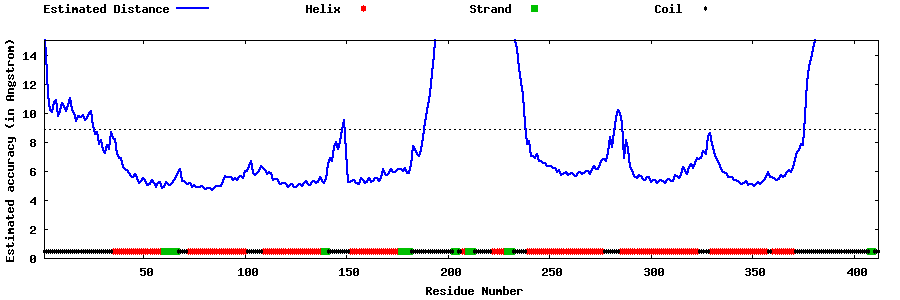 | |||
|
| |||
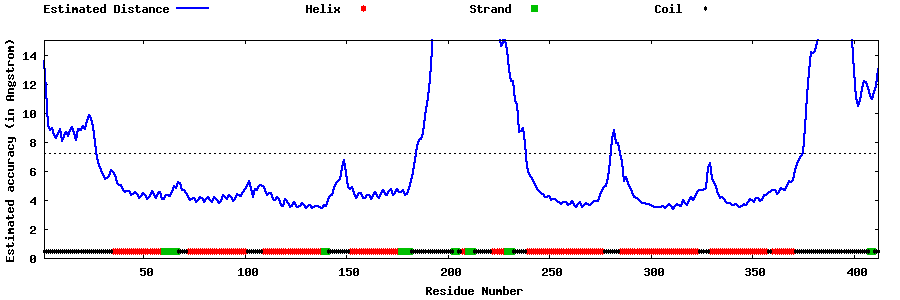 | |||
|
| |||