

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MDTGPDQSYFSGNHWFVFSVYLLTFLVGLPLNLLALVVFVGKLQRRPVAVDVLLLNLTASDLLLLLFLPFRMVEAANGMHWPLPFILCPLSGFIFFTTIYLTALFLAAVSIERFLSVAHPLWYKTRPRLGQAGLVSVACWLLASAHCSVVYVIEFSGDISHSQGTNGTCYLEFRKDQLAILLPVRLEMAVVLFVVPLIITSYCYSRLVWILGRGGSHRRQRRVAGLLAATLLNFLVCFGPYNVSHVVGYICGESPAWRIYVTLLSTLNSCVDPFVYYFSSSGFQADFHELLRRLCGLWGQWQQESSMELKEQKGGEEQRADRPAERKTSEHSQGCGTGGQVACAES | |
| CCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9999988777625530899999999999987898313533568888879999999999999999997599999996699999973879999999999999999999999998378866021113265624677777999999999998999997400012269983588567992378999999999999999999999999999999997257787545308999999999999996899999999999701038999999999999999899999308999999999998854665455667877777788998885789888877788899999987377779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MDTGPDQSYFSGNHWFVFSVYLLTFLVGLPLNLLALVVFVGKLQRRPVAVDVLLLNLTASDLLLLLFLPFRMVEAANGMHWPLPFILCPLSGFIFFTTIYLTALFLAAVSIERFLSVAHPLWYKTRPRLGQAGLVSVACWLLASAHCSVVYVIEFSGDISHSQGTNGTCYLEFRKDQLAILLPVRLEMAVVLFVVPLIITSYCYSRLVWILGRGGSHRRQRRVAGLLAATLLNFLVCFGPYNVSHVVGYICGESPAWRIYVTLLSTLNSCVDPFVYYFSSSGFQADFHELLRRLCGLWGQWQQESSMELKEQKGGEEQRADRPAERKTSEHSQGCGTGGQVACAES | |
| 7434344342403100002111200231232122001000030242310000000000100100000010100000344421001000110011123011100100000000000000000202423322100000010010011001010001102413447422100002034641212100111111301221120002000000010141654442100000000000001001231010100100033331010000000010001102010000440152014002310433453445434434455444444454445553454464444645443468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDTGPDQSYFSGNHWFVFSVYLLTFLVGLPLNLLALVVFVGKLQRRPVAVDVLLLNLTASDLLLLLFLPFRMVEAANGMHWPLPFILCPLSGFIFFTTIYLTALFLAAVSIERFLSVAHPLWYKTRPRLGQAGLVSVACWLLASAHCSVVYVIEFSGDISHSQGTNGTCYLEFRKDQLAILLPVRLEMAVVLFVVPLIITSYCYSRLVWILGRGGSHRRQRRVAGLLAATLLNFLVCFGPYNVSHVVGYICGESPAWRIYVTLLSTLNSCVDPFVYYFSSSGFQADFHELLRRLCGLWGQWQQESSMELKEQKGGEEQRADRPAERKTSEHSQGCGTGGQVACAES | |||||||||||||||||||||||||
| 1 | 5o9hA | 0.21 | 0.24 | 0.83 | 3.33 | Download | -------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAF--EAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEY--FPPKVLCGVDHD---KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETR-STKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR-------------------------------------------- | |||||||||||||||||||
| 2 | 5vblA | 0.22 | 0.24 | 0.83 | 3.94 | Download | ----CEYTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSSREKRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLRLRVSGAVATAVLWVLAALLAMPVMVLRTTGDL--ENTNKVQCYMDYSSSEWAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKEVEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLL------------------------------------------------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.22 | 0.19 | 0.84 | 3.54 | Download | -SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY-TKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.22 | 0.24 | 0.81 | 1.54 | Download | ---------SPAIPVIITAVYSVVFVVGLVGNSLVMFVII-RYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE--DVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.22 | 0.24 | 0.81 | 1.18 | Download | ---------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR-YTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE--DVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.23 | 0.19 | 0.84 | 3.36 | Download | -SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIV-RYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.22 | 0.24 | 0.80 | 1.73 | Download | ----------PAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY-TKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTK-V-REDVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------------------------------------------ | |||||||||||||||||||
| 8 | 4n6hA | 0.22 | 0.19 | 0.84 | 4.28 | Download | GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK-TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 9 | 5o9hA | 0.21 | 0.24 | 0.83 | 3.57 | Download | -------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAF--EAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVRE--EYFPPKVLCGVDHD---KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETR-STKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR-------------------------------------------- | |||||||||||||||||||
| 10 | 6b73A | 0.23 | 0.21 | 0.79 | 4.69 | Download | -----LGSISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKM-KTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKV-----RDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRSREKLRRITRLVLVVVAVFVVCWTPIHIFILVEALSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKR------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
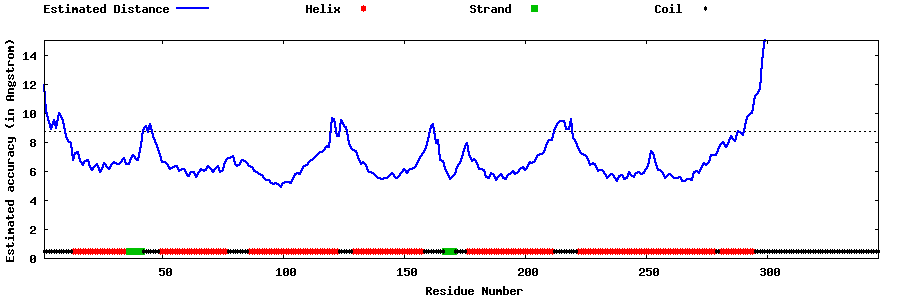
| Generated 3D models | Estimated local accuracy of models | ||
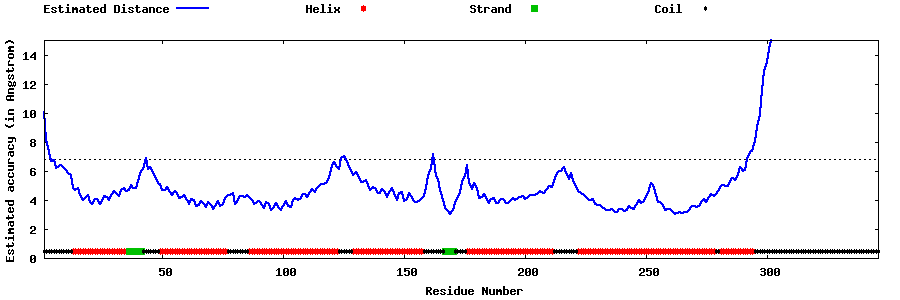 | |||
|
|
|||
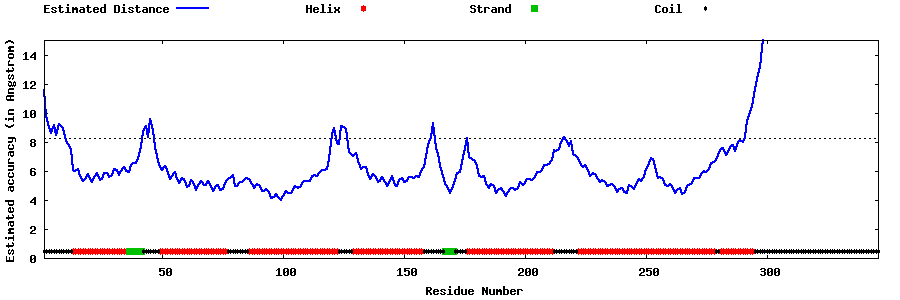 | |||
|
| |||
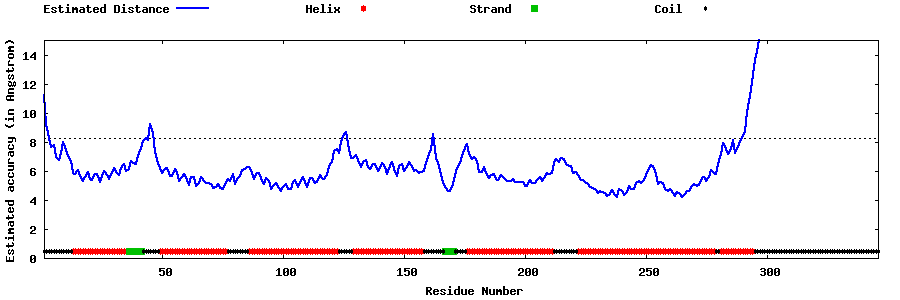 | |||
|
| |||
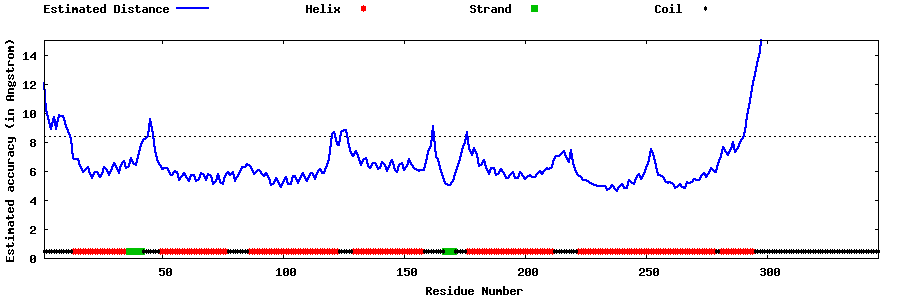 | |||
|
| |||
 | |||
|
| |||