

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MRAVFIQGAEEHPAAFCYQVNGSCPRTVHTLGIQLVIYLACAAGMLIIVLGNVFVAFAVSYFKALHTPTNFLLLSLALADMFLGLLVLPLSTIRSVESCWFFGDFLCRLHTYLDTLFCLTSIFHLCFISIDRHCAICDPLLYPSKFTVRVALRYILAGWGVPAAYTSLFLYTDVVETRLSQWLEEMPCVGSCQLLLNKFWGWLNFPLFFVPCLIMISLYVKIFVVATRQAQQITTLSKSLAGAAKHERKAAKTLGIAVGIYLLCWLPFTIDTMVDSLLHFITPPLVFDIFIWFAYFNSACNPIIYVFSYQWFRKALKLTLSQKVFSPQTRTVDLYQE | |
| CCCHHCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCSSCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHCCCCSSSSHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCC | |
| 9502208888885433665567889999885899999999999999999847872676872588883899999999999999999999999999983860177488869999999999999999999999833045433668653749999998999999999999999998110146677765577877674020011000021789999999999999999998462145422102455555653246437753799999986999999999997378771899999999999999599999996689999999999576779999997788889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MRAVFIQGAEEHPAAFCYQVNGSCPRTVHTLGIQLVIYLACAAGMLIIVLGNVFVAFAVSYFKALHTPTNFLLLSLALADMFLGLLVLPLSTIRSVESCWFFGDFLCRLHTYLDTLFCLTSIFHLCFISIDRHCAICDPLLYPSKFTVRVALRYILAGWGVPAAYTSLFLYTDVVETRLSQWLEEMPCVGSCQLLLNKFWGWLNFPLFFVPCLIMISLYVKIFVVATRQAQQITTLSKSLAGAAKHERKAAKTLGIAVGIYLLCWLPFTIDTMVDSLLHFITPPLVFDIFIWFAYFNSACNPIIYVFSYQWFRKALKLTLSQKVFSPQTRTVDLYQE | |
| 4543326346543331223212113444232001001111112112203323300000001033010000000000020221200000232001102320010210020000000000000130300000000000020031443123300000000012201210210010102333354233423113202023333103202321331020002002200200132033033334323333244331231000000000000211100000210152201300000000101230021000000105400300120001200344244242268 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCSSCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHCCCCSSSSHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCC MRAVFIQGAEEHPAAFCYQVNGSCPRTVHTLGIQLVIYLACAAGMLIIVLGNVFVAFAVSYFKALHTPTNFLLLSLALADMFLGLLVLPLSTIRSVESCWFFGDFLCRLHTYLDTLFCLTSIFHLCFISIDRHCAICDPLLYPSKFTVRVALRYILAGWGVPAAYTSLFLYTDVVETRLSQWLEEMPCVGSCQLLLNKFWGWLNFPLFFVPCLIMISLYVKIFVVATRQAQQITTLSKSLAGAAKHERKAAKTLGIAVGIYLLCWLPFTIDTMVDSLLHFITPPLVFDIFIWFAYFNSACNPIIYVFSYQWFRKALKLTLSQKVFSPQTRTVDLYQE | |||||||||||||||||||||||||
| 1 | 4iaqA | 0.33 | 0.31 | 0.82 | 2.82 | Download | ----------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF----------WRQASECVVNTD--HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSR------IIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI-----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-------------- | |||||||||||||||||||
| 2 | 4amjA | 0.36 | 0.33 | 0.86 | 4.39 | Download | -----------------------------SQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPGCCDFVTNRYAIASSIISFYIPLLIMIFVALRVYREAKEQIRKIDRKRKTSRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLLA---------------- | |||||||||||||||||||
| 3 | 3sn6R | 0.33 | 0.31 | 0.84 | 3.54 | Download | ------------------------------EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCYAE---ETCCDFFTNQYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRCL----KEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC---------------- | |||||||||||||||||||
| 4 | 3d4s | 0.33 | 0.31 | 0.85 | 1.55 | Download | --------------------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQHWYRATHQ----EAINCYTCCDFFTNQAYAISSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFDKARVDAYKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL--------------- | |||||||||||||||||||
| 5 | 3uon | 0.24 | 0.25 | 0.86 | 1.19 | Download | ------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVR-TVEDGECYIQFFS-NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGNAAKAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---------------- | |||||||||||||||||||
| 6 | 3sn6R | 0.33 | 0.31 | 0.84 | 3.07 | Download | -------------------------------VWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCYAEETCCDFF--TNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEG----RCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC---------------- | |||||||||||||||||||
| 7 | 3uon | 0.24 | 0.25 | 0.84 | 1.72 | Download | ---------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFVRTVEDGEC----YIQFFS-NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEGAAWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---------------- | |||||||||||||||||||
| 8 | 2rh1A | 0.32 | 0.31 | 0.85 | 4.04 | Download | ------------------------DE-----VWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYAEETCCDFFTNAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLN-----IFEMLRIDEKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCRS-PDFRIAFQELLCL--------------- | |||||||||||||||||||
| 9 | 4iaqA | 0.33 | 0.31 | 0.82 | 3.04 | Download | ----------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQASE----------CVVNTD--HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRII------QKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI-----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-------------- | |||||||||||||||||||
| 10 | 5jqhA | 0.30 | 0.31 | 0.93 | 3.50 | Download | LAKSYNQTPNRAKRVITTFRTGTWDAYAADEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYAEETCCDFFTNQAYAIASSISFYVPLVIMVFVYSRVFQEAKRQLD----------KFALKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCLR-------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
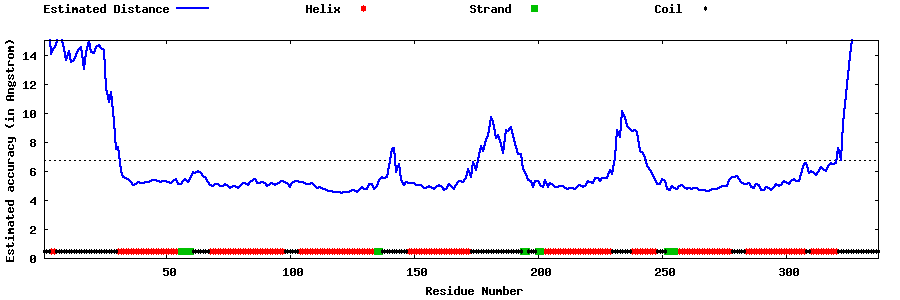
| Generated 3D models | Estimated local accuracy of models | ||
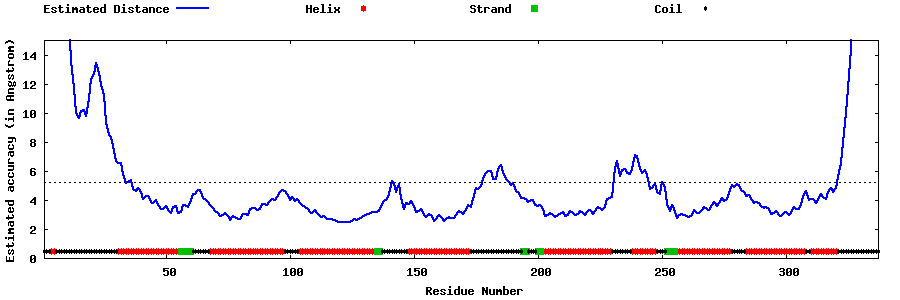 | |||
|
|
|||
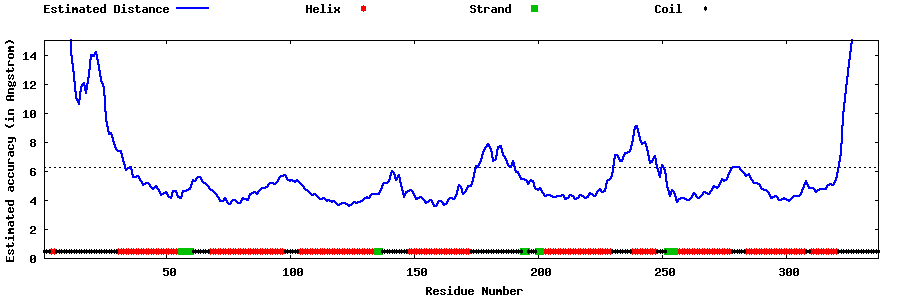 | |||
|
| |||
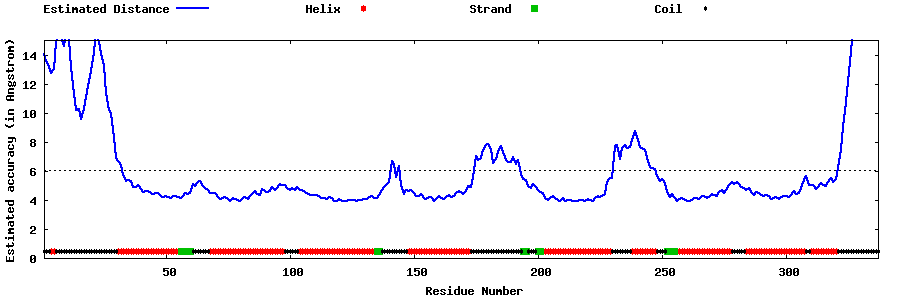 | |||
|
| |||
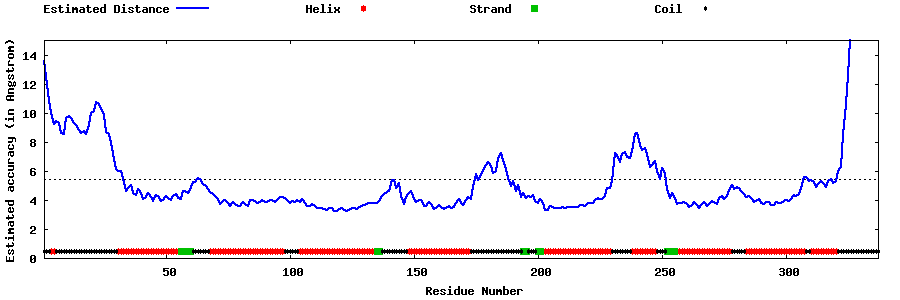 | |||
|
| |||
 | |||
|
| |||