

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MTNSSFFCPVYKDLEPFTYFFYLVFLVGIIGSCFATWAFIQKNTNHRCVSIYLINLLTADFLLTLALPVKIVVDLGVAPWKLKIFHCQVTACLIYINMYLSIIFLAFVSIDRCLQLTHSCKIYRIQEPGFAKMISTVVWLMVLLIMVPNMMIPIKDIKEKSNVGCMEFKKEFGRNWHLLTNFICVAIFLNFSAIILISNCLVIRQLYRNKDNENYPNVKKALINILLVTTGYIICFVPYHIVRIPYTLSQTEVITDCSTRISLFKAKEATLLLAVSNLCFDPILYYHLSKAFRSKVTETFASPKETKAQKEKLRCENNA | |
| CCCCCCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCC | |
| 9999999999766064879999999999998999889766337778699999999999999999943999999836799988866899999999999999999999999980477533101144666314777789999999999989999843365589837998459612279999999999752789999999999999999972877665533353799999999999998478999999999998567899269999999999999999999999899898228889999999846013466566888899998 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MTNSSFFCPVYKDLEPFTYFFYLVFLVGIIGSCFATWAFIQKNTNHRCVSIYLINLLTADFLLTLALPVKIVVDLGVAPWKLKIFHCQVTACLIYINMYLSIIFLAFVSIDRCLQLTHSCKIYRIQEPGFAKMISTVVWLMVLLIMVPNMMIPIKDIKEKSNVGCMEFKKEFGRNWHLLTNFICVAIFLNFSAIILISNCLVIRQLYRNKDNENYPNVKKALINILLVTTGYIICFVPYHIVRIPYTLSQTEVITDCSTRISLFKAKEATLLLAVSNLCFDPILYYHLSKAFRSKVTETFASPKETKAQKEKLRCENNA | |
| 7414344043354121001201200231333323001000123343210001013003011000000201000003444320120011000231331111102000001321300001003024233321000000001100000010100022143575331000102466322010011121312133312200200020011022145465344210000000000000120133023010010023132254041330021011101000130013200000000440141014003333445445553445668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCC MTNSSFFCPVYKDLEPFTYFFYLVFLVGIIGSCFATWAFIQKNTNHRCVSIYLINLLTADFLLTLALPVKIVVDLGVAPWKLKIFHCQVTACLIYINMYLSIIFLAFVSIDRCLQLTHSCKIYRIQEPGFAKMISTVVWLMVLLIMVPNMMIPIKDIKEKSNVGCMEFKKEFGRNWHLLTNFICVAIFLNFSAIILISNCLVIRQLYRNKDNENYPNVKKALINILLVTTGYIICFVPYHIVRIPYTLSQTEVITDCSTRISLFKAKEATLLLAVSNLCFDPILYYHLSKAFRSKVTETFASPKETKAQKEKLRCENNA | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.21 | 0.21 | 0.92 | 3.47 | Download | KIN-----VKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQYFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEK--KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 2 | 4xnwA | 0.23 | 0.25 | 0.91 | 4.42 | Download | -SFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIY----KEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFQAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRL----------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.21 | 0.21 | 0.91 | 3.94 | Download | ------PCQKINVKQLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE--KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 4 | 4djh | 0.22 | 0.27 | 0.88 | 1.53 | Download | ---------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSWDFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------- | |||||||||||||||||||
| 5 | 4xnv | 0.23 | 0.25 | 0.92 | 1.18 | Download | -SFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYP-KSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDEYRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKYEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFQAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR------------------------ | |||||||||||||||||||
| 6 | 4mbsA | 0.20 | 0.21 | 0.92 | 3.60 | Download | ----QKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE--KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 7 | 5t1a | 0.22 | 0.24 | 0.87 | 1.69 | Download | ----------------LPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKE--DSVYVCGPYFPRG-W-NNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIPPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.22 | 0.21 | 0.89 | 4.28 | Download | --------PCQKINVLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIF-TRSQKEGLHYTCSSHYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREKKRHRD------VRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 9 | 4xnvA | 0.24 | 0.24 | 0.91 | 3.57 | Download | -SFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPKSLGRLKK-KNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKEE----PLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFQTMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR------------------------ | |||||||||||||||||||
| 10 | 6b73A | 0.22 | 0.24 | 0.88 | 4.96 | Download | -----LGSISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTK-VRDVIECSLQFPDDDSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRSREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEAL-------GTSHSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKR------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
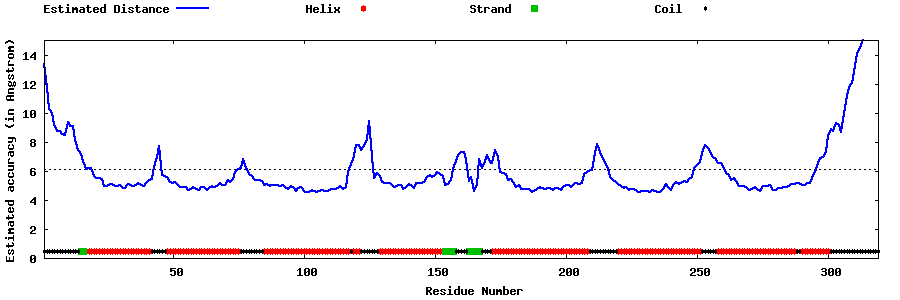
| Generated 3D models | Estimated local accuracy of models | ||
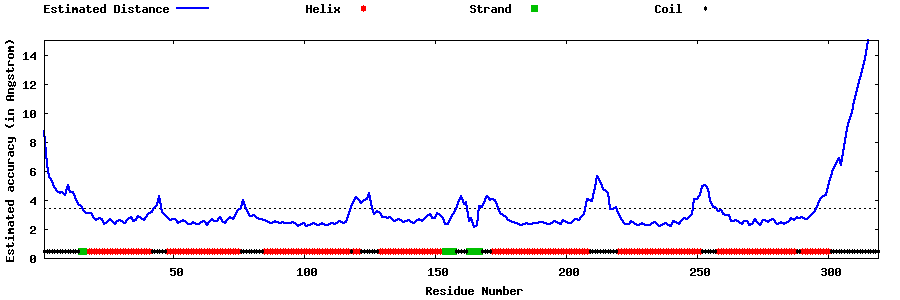 | |||
|
|
|||
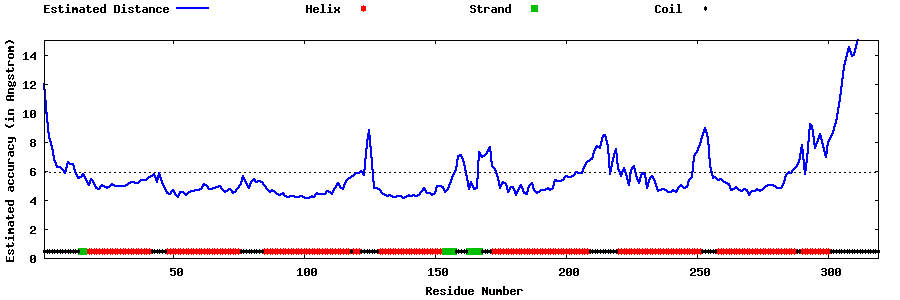 | |||
|
| |||
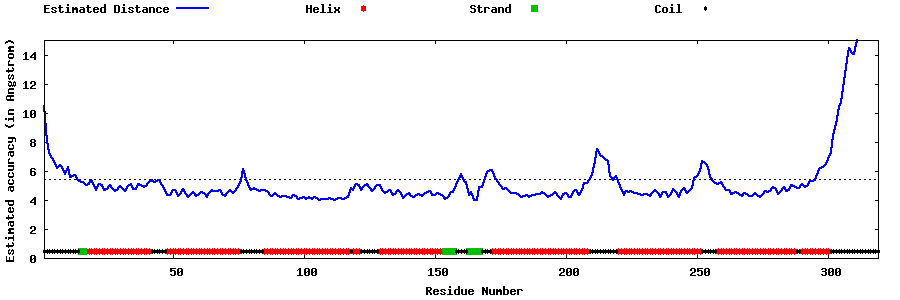 | |||
|
| |||
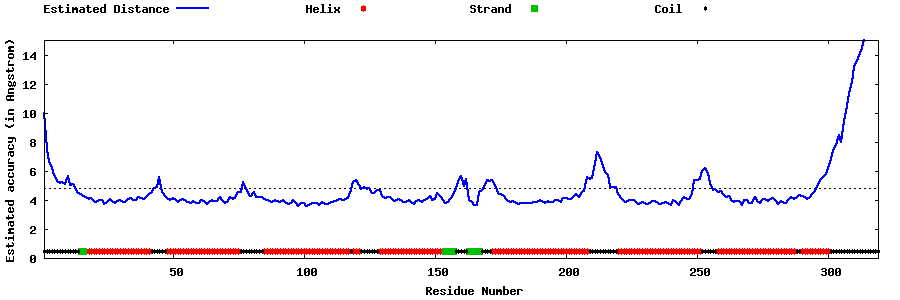 | |||
|
| |||
 | |||
|
| |||