

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MAPTEPWSPSPGSAPWDYSGLDGLEELELCPAGDLPYGYVYIPALYLAAFAVGLLGNAFVVWLLAGRRGPRRLVDTFVLHLAAADLGFVLTLPLWAAAAALGGRWPFGDGLCKLSSFALAGTRCAGALLLAGMSVDRYLAVVKLLEARPLRTPRCALASCCGVWAVALLAGLPSLVYRGLQPLPGGQDSQCGEEPSHAFQGLSLLLLLLTFVLPLVVTLFCYCRISRRLRRPPHVGRARRNSLRIIFAIESTFVGSWLPFSALRAVFHLARLGALPLPCPLLLALRWGLTIATCLAFVNSCANPLIYLLLDRSFRARALDGACGRTGRLARRISSASSLSRDDSSVFRCRAQAANTASASW | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCCCCCCCCCC | |
| 9999999998889888876778877666786655227786799999999999987899843525223467753356643799999999997299999997699876887899999999999999999999999998367875105203352436799998999999999998999962568808986799448808899999999999999999999999999999998267766522230778766777999999679999999999999548889817999999999999999999998999999951999999999981144567635788887887888764512579887778889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MAPTEPWSPSPGSAPWDYSGLDGLEELELCPAGDLPYGYVYIPALYLAAFAVGLLGNAFVVWLLAGRRGPRRLVDTFVLHLAAADLGFVLTLPLWAAAAALGGRWPFGDGLCKLSSFALAGTRCAGALLLAGMSVDRYLAVVKLLEARPLRTPRCALASCCGVWAVALLAGLPSLVYRGLQPLPGGQDSQCGEEPSHAFQGLSLLLLLLTFVLPLVVTLFCYCRISRRLRRPPHVGRARRNSLRIIFAIESTFVGSWLPFSALRAVFHLARLGALPLPCPLLLALRWGLTIATCLAFVNSCANPLIYLLLDRSFRARALDGACGRTGRLARRISSASSLSRDDSSVFRCRAQAANTASASW | |
| 8543442336643231113323424434115444240110000200120022023232100000012333210000000000102000000000000000334321002001210100132011000000000020000000000202422322100000000110000001000000202437643100001226302200111112303231220002000100110231554643220000000000000120313111010010023032133414222002001100100013000310000000044004201400242243234524444434454343343435446464444 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCCCCCCCCCC MAPTEPWSPSPGSAPWDYSGLDGLEELELCPAGDLPYGYVYIPALYLAAFAVGLLGNAFVVWLLAGRRGPRRLVDTFVLHLAAADLGFVLTLPLWAAAAALGGRWPFGDGLCKLSSFALAGTRCAGALLLAGMSVDRYLAVVKLLEARPLRTPRCALASCCGVWAVALLAGLPSLVYRGLQPLPGGQDSQCGEEPSHAFQGLSLLLLLLTFVLPLVVTLFCYCRISRRLRRPPHVGRARRNSLRIIFAIESTFVGSWLPFSALRAVFHLARLGALPLPCPLLLALRWGLTIATCLAFVNSCANPLIYLLLDRSFRARALDGACGRTGRLARRISSASSLSRDDSSVFRCRAQAANTASASW | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.26 | 0.23 | 0.81 | 3.20 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.26 | 0.24 | 0.81 | 3.91 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFF--------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.26 | 0.26 | 0.81 | 3.56 | Download | --------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD------PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.19 | 0.22 | 0.94 | 1.52 | Download | -DLRDNET-------WWYNPSIIVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVFPLMTISCF-LKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCN-CFDYISSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEW---------VTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCDDKETEAETEIPAGESSDAAPSADAAQMKE-- | |||||||||||||||||||
| 5 | 4djh | 0.26 | 0.23 | 0.77 | 1.15 | Download | ----------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVIECSLQFPDSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALG-----S-A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.26 | 0.23 | 0.81 | 3.28 | Download | ----------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ--WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.20 | 0.22 | 0.78 | 1.67 | Download | ----------------------------------PDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLE-GVLCNCSFDYRSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLE---W------VTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPW------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.27 | 0.23 | 0.80 | 4.75 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK----KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.26 | 0.23 | 0.81 | 3.39 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.24 | 0.22 | 0.80 | 3.89 | Download | -----------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKRRITRMVLVVVAVFIVCWTPIHIYVIIKAL-------ITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
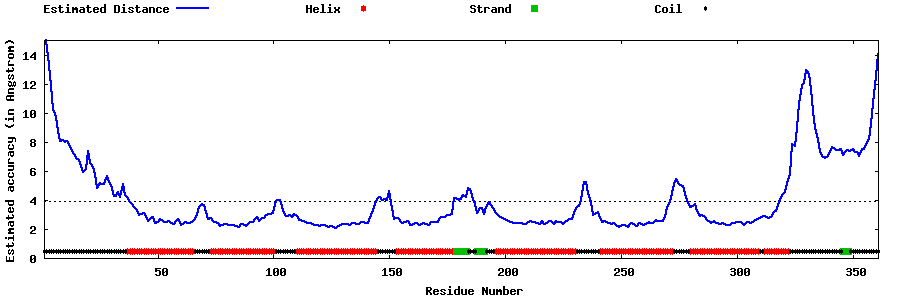 | |||
|
|
|||
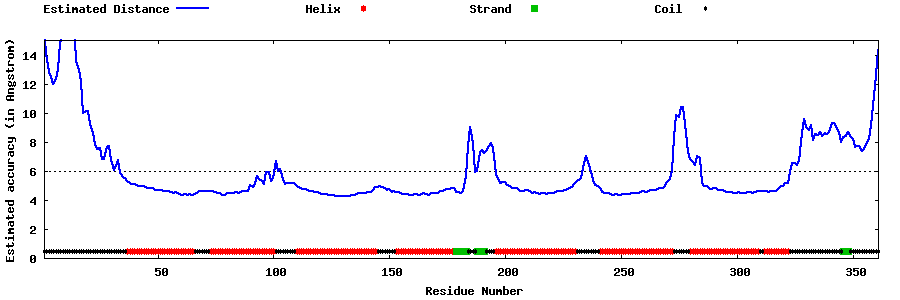 | |||
|
| |||
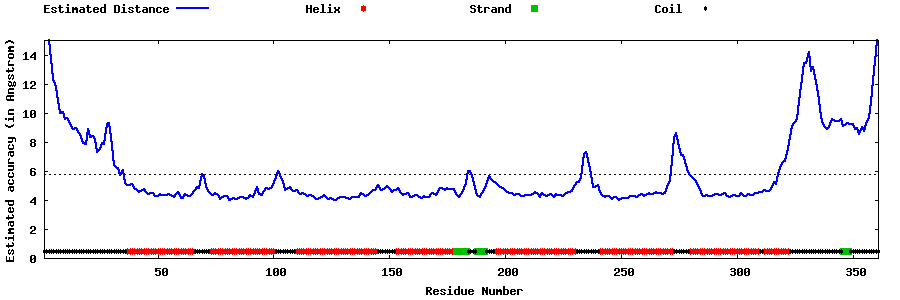 | |||
|
| |||
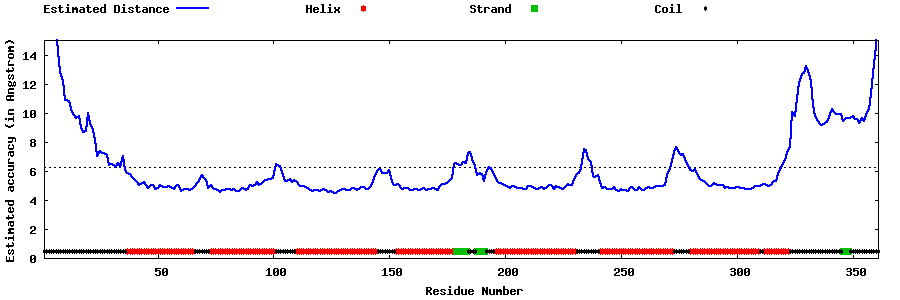 | |||
|
| |||
 | |||
|
| |||