

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDATACNESVDGSPVFYLLGIPSLPETFFLPVFFIFLLFYLLILMGNALILVAVVAEPSLHKPMYFFLINLSTLDILFTTTTVPKMLSLFLLGDRFLSFSSCLLQMYLFQSFTCSEAFILVVMAYDRYVAICHPLHYPVLMNPQTNATLAASAWLTALLLPIPAVVRTSQMAYNSIAYIYHCFCDHLAVVQASCSDTTPQTLMGFCIAMVVSFLPLLLVLLSYVHILASVLRISSLEGRAKAFSTCSSHLLVVGTYYSSIAIAYVAYRADLPLDFHIMGNVVYAILTPILNPLIYTLRNRDVKAAITKIMSQDPGCDRSI | |
| CCCCCCCCCCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSCHHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSHHCCCCCCCCHHHCCCCHHHHHHHHHHHHCCCCCCCCC | |
| 99763579884051314646999912527899999999999999989999999961887556599998889999874772489999898716997485999999999999999999999999987518870755457723587699999999999998999999999814168998913895678689987726464499999999999999987999999999999996207663776741166285798799985614326716898888988828998331305032456413561989999999982475531369 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDATACNESVDGSPVFYLLGIPSLPETFFLPVFFIFLLFYLLILMGNALILVAVVAEPSLHKPMYFFLINLSTLDILFTTTTVPKMLSLFLLGDRFLSFSSCLLQMYLFQSFTCSEAFILVVMAYDRYVAICHPLHYPVLMNPQTNATLAASAWLTALLLPIPAVVRTSQMAYNSIAYIYHCFCDHLAVVQASCSDTTPQTLMGFCIAMVVSFLPLLLVLLSYVHILASVLRISSLEGRAKAFSTCSSHLLVVGTYYSSIAIAYVAYRADLPLDFHIMGNVVYAILTPILNPLIYTLRNRDVKAAITKIMSQDPGCDRSI | |
| 65434465130200100000006305212020112123212312332320010010013000000010001002012010020030020011433403020020011001200320000000002100000010020002001200000012003100310300010000020037240100001232003000210220001022213331331231023011100000010214612210000000000000011200100001021333443320000000232133003000020310130023003334325665 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSCHHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSHHCCCCCCCCHHHCCCCHHHHHHHHHHHHCCCCCCCCC MDATACNESVDGSPVFYLLGIPSLPETFFLPVFFIFLLFYLLILMGNALILVAVVAEPSLHKPMYFFLINLSTLDILFTTTTVPKMLSLFLLGDRFLSFSSCLLQMYLFQSFTCSEAFILVVMAYDRYVAICHPLHYPVLMNPQTNATLAASAWLTALLLPIPAVVRTSQMAYNSIAYIYHCFCDHLAVVQASCSDTTPQTLMGFCIAMVVSFLPLLLVLLSYVHILASVLRISSLEGRAKAFSTCSSHLLVVGTYYSSIAIAYVAYRADLPLDFHIMGNVVYAILTPILNPLIYTLRNRDVKAAITKIMSQDPGCDRSI | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.22 | 0.87 | 3.53 | Download | -------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFT-FFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.23 | 0.88 | 2.15 | Download | -----RGENFMDIECFMVLN-----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFID-FHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 3 | 4iaqA | 0.18 | 0.22 | 0.85 | 2.17 | Download | -----------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLP--------PFFWRQASE-------------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQKYLLMERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.21 | 0.88 | 1.53 | Download | -----------------------SPAI-PVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR-E--DVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------- | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.19 | 0.90 | 1.24 | Download | AEQLKTTRN-AYIQKYLILNSSDCPYIFV-MIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVF-F------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL---------- | |||||||||||||||||||
| 6 | 3emlA | 0.19 | 0.22 | 0.87 | 3.75 | Download | --------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 7 | 4iaq | 0.21 | 0.22 | 0.83 | 1.71 | Download | -----------------------ISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQAS----------ECVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 8 | 2z73A | 0.16 | 0.22 | 0.95 | 2.92 | Download | -ETW--WYNPSIVVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWIGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFG------WGAYTLEGVLCD-------YISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFD | |||||||||||||||||||
| 9 | 3emlA | 0.19 | 0.22 | 0.87 | 4.89 | Download | -------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.21 | 0.91 | 5.52 | Download | ----------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEP | |||||||||||||||||||
| ||||||||||||||||||||||||||
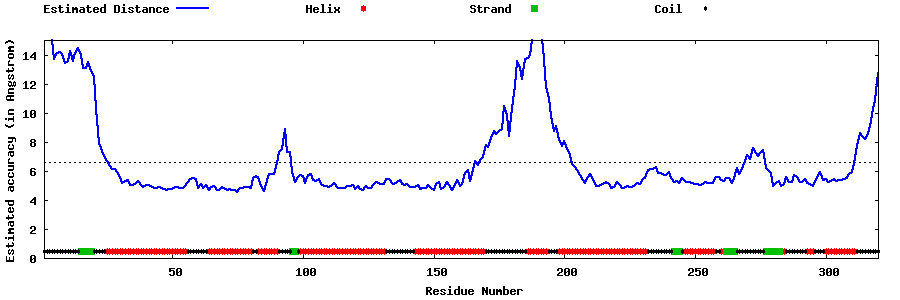
| Generated 3D models | Estimated local accuracy of models | ||
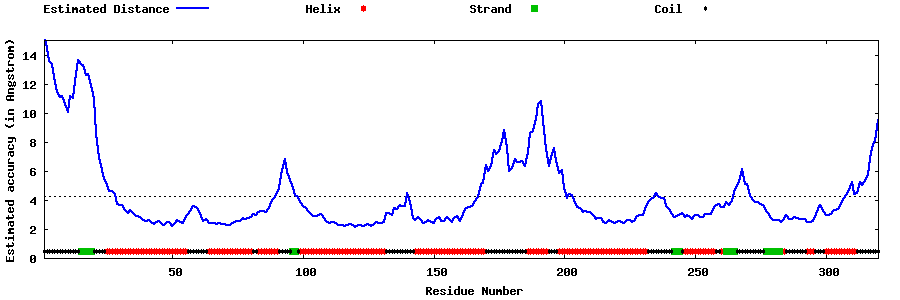 | |||
|
|
|||
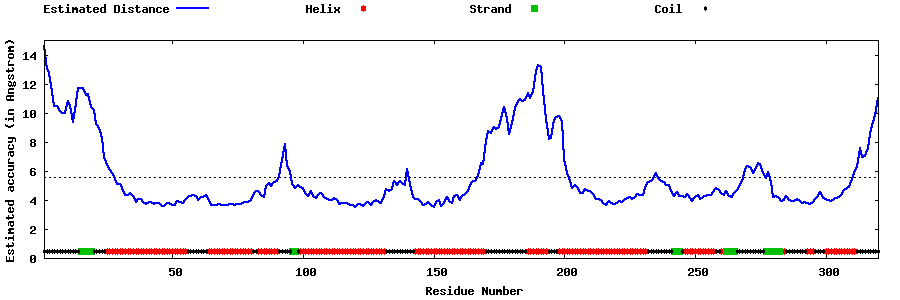 | |||
|
| |||
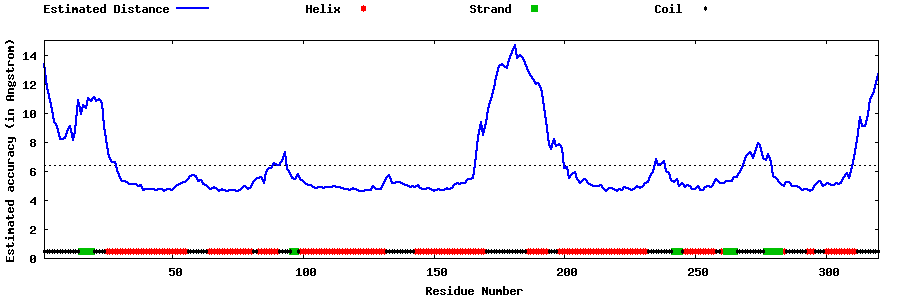 | |||
|
| |||
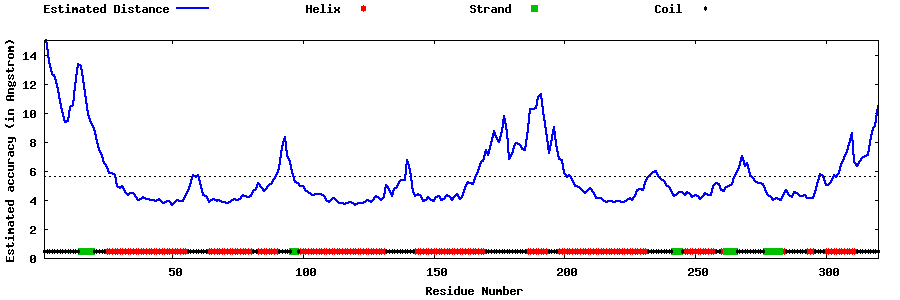 | |||
|
| |||
 | |||
|
| |||