

You can:
| Name | CHEMBL1632411 |
|---|---|
| Molecular formula | C15H19N3O |
| IUPAC name | (NZ)-N-[1H-indol-2-yl-(1-methylpiperidin-4-yl)methylidene]hydroxylamine |
| Molecular weight | 257.337 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 2.8 |
| Synonyms | N/A |
| Inchi Key | MQAUVLYNYCPXHD-ICFOKQHNSA-N |
| Inchi ID | InChI=1S/C15H19N3O/c1-18-8-6-11(7-9-18)15(17-19)14-10-12-4-2-3-5-13(12)16-14/h2-5,10-11,16,19H,6-9H2,1H3/b17-15- |
| PubChem CID | 136192886 |
| ChEMBL | CHEMBL1632411 |
| IUPHAR | N/A |
| BindingDB | N/A |
| DrugBank | N/A |
Structure | 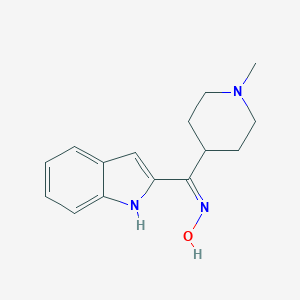 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
You can:
| GLASS ID | Name | UniProt | Gene | Species | Length |
|---|---|---|---|---|---|
| 563984 | Histamine H1 receptor | P35367 | HRH1 | Homo sapiens (Human) | 487 |
| 563986 | Histamine H2 receptor | P25021 | HRH2 | Homo sapiens (Human) | 359 |
| 563983 | Histamine H3 receptor | Q9Y5N1 | HRH3 | Homo sapiens (Human) | 445 |
| 563985 | Histamine H4 receptor | Q91ZY1 | Hrh4 | Rattus norvegicus (Rat) | 391 |
| 563987 | Histamine H4 receptor | Q9H3N8 | HRH4 | Homo sapiens (Human) | 390 |
| 563988 | Histamine H4 receptor | Q91ZY2 | Hrh4 | Mus musculus (Mouse) | 391 |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417