

You can:
| Name | Metabotropic glutamate receptor 1 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Grm1 |
| Synonym | mGluR1 SCAR13 mGlu1 receptor metabotropic glutamate receptor 1 wobl [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 1199 |
| Amino acid sequence | MVRLLLIFFPMIFLEMSILPRMPDRKVLLAGASSQRSVARMDGDVIIGALFSVHHQPPAEKVPERKCGEIREQYGIQRVEAMFHTLDKINADPVLLPNITLGSEIRDSCWHSSVALEQSIEFIRDSLISIRDEKDGLNRCLPDGQTLPPGRTKKPIAGVIGPGSSSVAIQVQNLLQLFDIPQIAYSATSIDLSDKTLYKYFLRVVPSDTLQARAMLDIVKRYNWTYVSAVHTEGNYGESGMDAFKELAAQEGLCIAHSDKIYSNAGEKSFDRLLRKLRERLPKARVVVCFCEGMTVRGLLSAMRRLGVVGEFSLIGSDGWADRDEVIEGYEVEANGGITIKLQSPEVRSFDDYFLKLRLDTNTRNPWFPEFWQHRFQCRLPGHLLENPNFKKVCTGNESLEENYVQDSKMGFVINAIYAMAHGLQNMHHALCPGHVGLCDAMKPIDGRKLLDFLIKSSFVGVSGEEVWFDEKGDAPGRYDIMNLQYTEANRYDYVHVGTWHEGVLNIDDYKIQMNKSGMVRSVCSEPCLKGQIKVIRKGEVSCCWICTACKENEFVQDEFTCRACDLGWWPNAELTGCEPIPVRYLEWSDIESIIAIAFSCLGILVTLFVTLIFVLYRDTPVVKSSSRELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILAGSKKKICTRKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPMPILSYPSIKEVYLICNTSNLGVVAPVGYNGLLIMSCTYYAFKTRNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYKIITTCFAVSLSVTVALGCMFTPKMYIIIAKPERNVRSAFTTSDVVRMHVGDGKLPCRSNTFLNIFRRKKPGAGNANSNGKSVSWSEPGGRQAPKGQHVWQRLSVHVKTNETACNQTAVIKPLTKSYQGSGKSLTFSDASTKTLYNVEEEDNTPSAHFSPPSSPSMVVHRRGPPVATTPPLPPHLTAEETPLFLADSVIPKGLPPPLPQQQPQQPPPQQPPQQPKSLMDQLQGVVTNFGSGIPDFHAVLAGPGTPGNSLRSLYPPPPPPQHLQMLPLHLSTFQEESISPPGEDIDDDSERFKLLQEFVYEREGNTEEDELEEEEDLPTASKLTPEDSPALTPPSPFRDSVASGSSVPSSPVSESVLCTPPNVTYASVILRDYKQSSSTL |
| UniProt | P23385 |
| Protein Data Bank | 1iss, 1isr, 1ewk |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 1iss. |
| BioLiP | BL0023849, BL0023850,BL0023851, BL0011887,BL0011889, BL0011888,BL0011890 |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4477 |
| IUPHAR | 289 |
| DrugBank | N/A |
| Name | CHEMBL238262 |
|---|---|
| Molecular formula | C17H11N5OS |
| IUPAC name | 13-(4-methylphenyl)-10-thia-3,4,8,13,15-pentazatetracyclo[7.7.0.02,6.011,16]hexadeca-1,4,6,8,11(16),14-hexaen-12-one |
| Molecular weight | 333.369 |
| Hydrogen bond acceptor | 5 |
| Hydrogen bond donor | 1 |
| XlogP | 3.1 |
| Synonyms | BDBM50224403 SCHEMBL8243816 8-(4-methylphenyl)-1H-pyrazolo[3'''',4'''':4'',5'']pyrido[3'',2'':4,5]thieno[3,2-d]pyrimidin-7(8H)-one |
| Inchi Key | AJSOPCTYDMXEJW-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C17H11N5OS/c1-9-2-4-11(5-3-9)22-8-19-14-12-13-10(7-20-21-13)6-18-16(12)24-15(14)17(22)23/h2-8H,1H3,(H,20,21) |
| PubChem CID | 16038352 |
| ChEMBL | CHEMBL238262 |
| IUPHAR | N/A |
| BindingDB | 50224403 |
| DrugBank | N/A |
Structure | 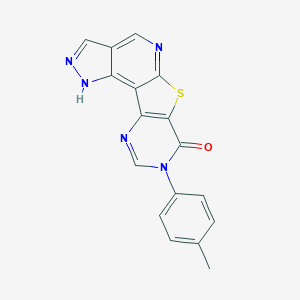 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | 17.8 nM | PMID17929793 | ChEMBL |
| Ki | 18.0 nM | PMID17929793 | BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417