

You can:
| Name | Mu-type opioid receptor |
|---|---|
| Species | Cavia porcellus (Guinea pig) |
| Gene | OPRM1 |
| Synonym | M-OR-1 MOR-1 |
| Disease | N/A for non-human GPCRs |
| Length | 98 |
| Amino acid sequence | YTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGTILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKTVNVCNWI |
| UniProt | P97266 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4354 |
| IUPHAR | N/A |
| DrugBank | N/A |
| Name | morphine |
|---|---|
| Molecular formula | C17H19NO3 |
| IUPAC name | (4R,4aR,7S,7aR,12bS)-3-methyl-2,4,4a,7,7a,13-hexahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline-7,9-diol |
| Molecular weight | 285.343 |
| Hydrogen bond acceptor | 4 |
| Hydrogen bond donor | 2 |
| XlogP | 0.8 |
| Synonyms | (-)(5.alpha.,6.alpha.)-7,8-Didehydro-4,5-epoxy-17-methylmorphinan-3,6-diol Duromorph (5alpha,6alpha)-17-methyl-7,8-didehydro-4,5-epoxymorphinan-3,6-diol Infumorph SDZ202-250 [ Show all ] |
| Inchi Key | BQJCRHHNABKAKU-KBQPJGBKSA-N |
| Inchi ID | InChI=1S/C17H19NO3/c1-18-7-6-17-10-3-5-13(20)16(17)21-15-12(19)4-2-9(14(15)17)8-11(10)18/h2-5,10-11,13,16,19-20H,6-8H2,1H3/t10-,11+,13-,16-,17-/m0/s1 |
| PubChem CID | 5288826 |
| ChEMBL | CHEMBL70 |
| IUPHAR | 1627 |
| BindingDB | 50000092 |
| DrugBank | DB00295 |
Structure | 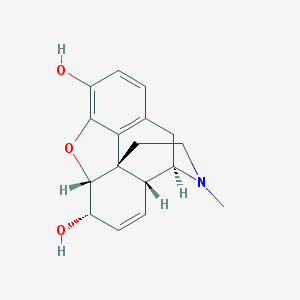 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Dose ratio | 45.0 - | PMID1310115 | ChEMBL |
| IC50 | 4.0 nM | PMID2887656 | BindingDB,ChEMBL |
| IC50 | 8.5 nM | , Bioorg. Med. Chem. Lett., (1994) 4:21:2527 | BindingDB,ChEMBL |
| IC50 | 29.0 nM | PMID25129170 | BindingDB |
| IC50 | 29.3 nM | PMID25129170 | ChEMBL |
| IC50 | 38.0 nM | PMID1851843 | BindingDB,ChEMBL |
| IC50 | 57.3 nM | PMID7154002 | BindingDB,ChEMBL |
| IC50 | 58.8 nM | PMID12166947 | ChEMBL |
| IC50 | 59.0 nM | PMID12166947 | BindingDB |
| IC50 | 70.0 nM | PMID2828622, PMID6276540 | BindingDB,ChEMBL |
| IC50 | 71.6 nM | PMID1967312 | ChEMBL |
| IC50 | 72.0 nM | PMID1967312 | BindingDB |
| IC50 | 77.0 nM | PMID6313921 | BindingDB,ChEMBL |
| IC50 | 311.0 nM | PMID15857143 | BindingDB,ChEMBL |
| IC50 | 29300.0 nM | PMID18370374 | BindingDB,ChEMBL |
| Ki | 0.88 nM | PMID19027293, PMID11000010, PMID11425545 | BindingDB |
| Ki | 0.88 nM | PMID11425545, PMID11000010, PMID19027293 | ChEMBL |
| Ki | 1.0 nM | PMID11121615 | BindingDB |
| Ki | 1.1 nM | PMID21641798 | BindingDB,ChEMBL |
| Ki | 1.8 nM | PMID2832603 | BindingDB,ChEMBL |
| Ki | 1.98 nM | PMID21641219 | BindingDB,ChEMBL |
| Ki | 2.0 nM | PMID9686407 | BindingDB |
| Ki | 2.2 nM | PMID21481987 | BindingDB,ChEMBL |
| Ki | 2.5 nM | PMID16777416 | BindingDB,ChEMBL |
| Ki | 2.69 nM | PMID6248635 | BindingDB |
| Ki | 3.0 nM | PMID12930147 | BindingDB,ChEMBL |
| Ki | 3.3 nM | PMID1652025, PMID1846921, PMID1323679 | BindingDB,ChEMBL |
| Ki | 3.467 nM | PMID11960505 | ChEMBL |
| Ki | 3.5 nM | PMID23517479 | ChEMBL |
| Ki | 3.9 nM | PMID26411794 | BindingDB,ChEMBL |
| Ki | 6.5 nM | PMID9873603 | BindingDB |
| Ki | 6.51 nM | PMID9873603 | ChEMBL |
| Ki | 7.7 nM | PMID1967312 | BindingDB |
| Ki | 7.71 nM | PMID1967312 | ChEMBL |
| Ki | 38.0 nM | PMID7932535, PMID9873439 | BindingDB,ChEMBL |
| Relative affinity | 98.5 % | PMID11960505 | ChEMBL |
| Relative Potency | 1.0 - | PMID6313921 | ChEMBL |
| Relative potency | 1.0 - | PMID2887656 | ChEMBL |
| Relative potency | 100.0 - | PMID7154002 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417