

You can:
| Name | 5-hydroxytryptamine receptor 1A |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Htr1a |
| Synonym | 5-HT1A receptor 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled 5-HT1A ADRB2RL1 ADRBRL1 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 422 |
| Amino acid sequence | MDVFSFGQGNNTTASQEPFGTGGNVTSISDVTFSYQVITSLLLGTLIFCAVLGNACVVAAIALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPEDRSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVRKVEKKGAGTSLGTSSAPPPKKSLNGQPGSGDWRRCAENRAVGTPCTNGAVRQGDDEATLEVIEVHRVGNSKEHLPLPSESGSNSYAPACLERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHMPALLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFCRR |
| UniProt | P19327 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL273 |
| IUPHAR | 1 |
| DrugBank | N/A |
| Name | buspirone |
|---|---|
| Molecular formula | C21H31N5O2 |
| IUPAC name | 8-[4-(4-pyrimidin-2-ylpiperazin-1-yl)butyl]-8-azaspiro[4.5]decane-7,9-dione |
| Molecular weight | 385.512 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 0 |
| XlogP | 2.6 |
| Synonyms | ZINC1530571 DTXSID2022707 8-[4-(4-pyrimidin-2-ylpiperazin-1-yl)butyl]-8-azaspiro[4.5]decane-7,9-dione KB-250161 8-{4-[4-(pyrimidin-2-yl)piperazin-1-yl]butyl}-8-azaspiro[4.5]decane-7,9-dione [ Show all ] |
| Inchi Key | QWCRAEMEVRGPNT-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C21H31N5O2/c27-18-16-21(6-1-2-7-21)17-19(28)26(18)11-4-3-10-24-12-14-25(15-13-24)20-22-8-5-9-23-20/h5,8-9H,1-4,6-7,10-17H2 |
| PubChem CID | 2477 |
| ChEMBL | CHEMBL49 |
| IUPHAR | 36 |
| BindingDB | 50001859 |
| DrugBank | DB00490 |
Structure | 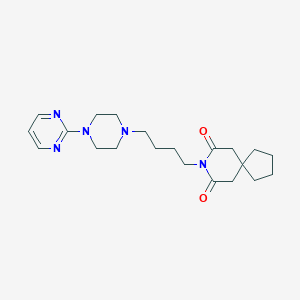 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Hill coefficient | 0.95 - | PMID3172131 | ChEMBL |
| IC50 | 7.1 nM | N/A | BindingDB |
| IC50 | 7.13 nM | Bioorg. Med. Chem. Lett., (1997) 7:22:2857 | ChEMBL |
| IC50 | 11.0 nM | PMID15914001, PMID11229779 | BindingDB,ChEMBL |
| IC50 | 12.8 nM | PMID7699710, PMID8289207 | BindingDB,ChEMBL |
| IC50 | 16.0 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| IC50 | 21.88 nM | PMID3373482 | ChEMBL |
| IC50 | 22.0 nM | PMID3373482 | BindingDB |
| IC50 | 24.0 nM | PMID8496920 | BindingDB,ChEMBL |
| IC50 | 30.0 nM | PMID8960552, PMID8759642 | BindingDB,ChEMBL |
| IC50 | 50.0 nM | PMID9986719 | BindingDB,ChEMBL |
| IC50 | 52.0 nM | PMID9083484 | BindingDB,ChEMBL |
| IC50 | 60.0 nM | PMID8863806, PMID7912735 | BindingDB,ChEMBL |
| IC50 | 60.0 nM | PMID8863806 | BindingDB |
| Inhibition | 43.0 % | PMID8831769 | ChEMBL |
| Inhibition | 64.0 % | PMID8831769 | ChEMBL |
| Inhibition | 73.0 % | PMID8831769 | ChEMBL |
| Inhibition | 75.0 % | PMID8831769 | ChEMBL |
| Inhibition | 84.0 % | PMID8831769 | ChEMBL |
| Ki | 3.8 nM | PMID18486277 | BindingDB,ChEMBL |
| Ki | 5.0 nM | PMID1346653, PMID7473547 | BindingDB,ChEMBL |
| Ki | 7.0 nM | PMID9083484 | BindingDB,ChEMBL |
| Ki | 9.3 nM | PMID9083484 | BindingDB,ChEMBL |
| Ki | 9.307 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Ki | 10.0 nM | PMID10602693, PMID2565399, PMID2898533 | BindingDB,ChEMBL |
| Ki | 12.3 nM | PMID8676348 | BindingDB,ChEMBL |
| Ki | 12.58 nM | PMID10611634 | BindingDB |
| Ki | 13.8 nM | PMID7752194 | ChEMBL |
| Ki | 14.0 nM | PMID7752194 | BindingDB |
| Ki | 14.79 nM | PMID8289207 | ChEMBL |
| Ki | 15.0 nM | PMID8831769, PMID3172131, PMID7902439, PMID18598015 | BindingDB,ChEMBL |
| Ki | 15.4 nM | PMID7731013 | BindingDB,ChEMBL |
| Ki | 19.95 nM | PMID8568799 | ChEMBL |
| Ki | 20.0 nM | PMID25435254 | BindingDB,ChEMBL |
| Ki | 20.5 nM | PMID8893838, Bioorg. Med. Chem. Lett., (1996) 6:6:689 | BindingDB,ChEMBL |
| Ki | 21.0 nM | N/A | BindingDB |
| Ki | 29.51 nM | PMID6225026 | BindingDB |
| Ki | 32.0 nM | PMID11728188 | BindingDB,ChEMBL |
| Ki | 36.0 nM | PMID18603331 | BindingDB,ChEMBL |
| Ki | 46.0 nM | PMID8289183 | BindingDB |
| Ki | 46.1 nM | PMID8289183 | ChEMBL |
| Ki | 47.86 nM | PMID8584042 | BindingDB |
| Ki | 56.23 nM | PMID8584042 | BindingDB |
| Ki | 68.7 nM | PMID8398139 | BindingDB |
| Ki | 891.25 nM | PMID6225026 | BindingDB |
| Ks | 16.0 nM | PMID21232965 | ChEMBL |
| Ratio | 5.0 - | PMID11229779 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417