

You can:
| Name | D(2) dopamine receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Drd2 |
| Synonym | D2 receptor D2(415) and D2(444) D2A and D2B D2R Dopamine D2 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 444 |
| Amino acid sequence | MDPLNLSWYDDDLERQNWSRPFNGSEGKADRPHYNYYAMLLTLLIFIIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMIAIVWVLSFTISCPLLFGLNNTDQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRKRRKRVNTKRSSRAFRANLKTPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRMDAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSNPDSPAKPEKNGHAKIVNPRIAKFFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFMKILHC |
| UniProt | P61169 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL339 |
| IUPHAR | 215 |
| DrugBank | N/A |
| Name | sulpiride |
|---|---|
| Molecular formula | C15H23N3O4S |
| IUPAC name | N-[(1-ethylpyrrolidin-2-yl)methyl]-2-methoxy-5-sulfamoylbenzamide |
| Molecular weight | 341.426 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 2 |
| XlogP | 0.6 |
| Synonyms | Ozoderpin Pyrikappl S0501 SR-01000075402-2 Sulpirida [INN-Spanish] [ Show all ] |
| Inchi Key | BGRJTUBHPOOWDU-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C15H23N3O4S/c1-3-18-8-4-5-11(18)10-17-15(19)13-9-12(23(16,20)21)6-7-14(13)22-2/h6-7,9,11H,3-5,8,10H2,1-2H3,(H,17,19)(H2,16,20,21) |
| PubChem CID | 5355 |
| ChEMBL | CHEMBL26 |
| IUPHAR | 5501 |
| BindingDB | 11638 |
| DrugBank | DB00391 |
Structure | 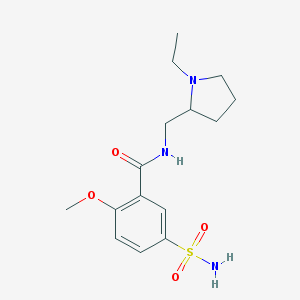 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 33.0 nM | PMID3172140 | BindingDB,ChEMBL |
| IC50 | 36.0 nM | PMID3172140 | BindingDB,ChEMBL |
| IC50 | 210.0 nM | PMID7901417 | BindingDB,ChEMBL |
| IC50 | 233.0 nM | PMID4040977 | BindingDB,ChEMBL |
| Kd | 1.5 nM | PMID2966245 | BindingDB,ChEMBL |
| Ki | 0.21 nM | PMID10327430 | BindingDB |
| Ki | 2.5 nM | PMID11171942 | BindingDB |
| Ki | 5.51 nM | PMID2522993 | BindingDB,ChEMBL |
| Ki | 6.58 nM | PMID2531826 | BindingDB |
| Ki | 8.2 nM | PMID8278895 | BindingDB |
| Ki | 9.2 nM | PMID1975644 | BindingDB |
| Ki | 10.2 nM | PMID8278895 | BindingDB |
| Ki | 10.5 nM | PMID8278895 | BindingDB |
| Ki | 23.0 nM | PMID2138666, PMID3828800 | BindingDB |
| Ki | 24.0 nM | PMID2958621 | BindingDB |
| Ki | 42.0 nM | PMID2138666 | BindingDB |
| Ki | 57.0 nM | PMID8301592 | BindingDB |
| Ki | 70.0 nM | PMID2958621 | BindingDB |
| Ki | 71.6 nM | PMID8301592 | BindingDB |
| Ki | 78.0 nM | PMID10327430 | BindingDB |
| Ki | 85.0 nM | PMID1975644 | BindingDB |
| Ki | 122.0 nM | PMID2869639 | BindingDB |
| Ki | 150.0 nM | PMID3130534 | BindingDB |
| Ki | 160.0 nM | PMID2532362 | BindingDB |
| Ki | 180.0 nM | PMID6655559 | BindingDB |
| Ki | 205.0 nM | PMID2532362 | BindingDB |
| Ki | 233.0 nM | PMID2869639 | BindingDB |
| Ki | 240.0 nM | PMID9748351 | BindingDB,ChEMBL |
| Ki | 320.0 nM | PMID6655559 | BindingDB |
| Ki | 340.0 nM | PMID10327430 | BindingDB |
| Ki | 518.0 nM | PMID3828800 | BindingDB |
| Ki | 636.0 nM | PMID1356154 | BindingDB |
| Ki | 730.0 nM | PMID2869639 | BindingDB |
| Ki | 1102.0 nM | PMID6387355 | BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417