

You can:
| Name | 5-hydroxytryptamine receptor 2C |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR2C |
| Synonym | Serotonin receptor 2C serotonin 1c receptor 5-HT1C 5-HT2C 5-HT-2C [ Show all ] |
| Disease | Pain Sleep initiation and maintenance disorders; Primary insomnia; Schizophrenia Unspecified Depression Drug abuse [ Show all ] |
| Length | 458 |
| Amino acid sequence | MVNLRNAVHSFLVHLIGLLVWQCDISVSPVAAIVTDIFNTSDGGRFKFPDGVQNWPALSIVIIIIMTIGGNILVIMAVSMEKKLHNATNYFLMSLAIADMLVGLLVMPLSLLAILYDYVWPLPRYLCPVWISLDVLFSTASIMHLCAISLDRYVAIRNPIEHSRFNSRTKAIMKIAIVWAISIGVSVPIPVIGLRDEEKVFVNNTTCVLNDPNFVLIGSFVAFFIPLTIMVITYCLTIYVLRRQALMLLHGHTEEPPGLSLDFLKCCKRNTAEEENSANPNQDQNARRRKKKERRPRGTMQAINNERKASKVLGIVFFVFLIMWCPFFITNILSVLCEKSCNQKLMEKLLNVFVWIGYVCSGINPLVYTLFNKIYRRAFSNYLRCNYKVEKKPPVRQIPRVAATALSGRELNVNIYRHTNEPVIEKASDNEPGIEMQVENLELPVNPSSVVSERISSV |
| UniProt | P28335 |
| Protein Data Bank | 6bqg, 6bqh |
| GPCR-HGmod model | P28335 |
| 3D structure model | This structure is from PDB ID 6bqg. |
| BioLiP | BL0404805, BL0404806 |
| Therapeutic Target Database | T83813 |
| ChEMBL | CHEMBL225 |
| IUPHAR | 8 |
| DrugBank | BE0004957, BE0004881, BE0000533 |
| Name | ritanserin |
|---|---|
| Molecular formula | C27H25F2N3OS |
| IUPAC name | 6-[2-[4-[bis(4-fluorophenyl)methylidene]piperidin-1-yl]ethyl]-7-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one |
| Molecular weight | 477.574 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 0 |
| XlogP | 5.2 |
| Synonyms | NCGC00015877-07 AB00053288_14 NCGC00261768-01 B6898 R-55667 [ Show all ] |
| Inchi Key | JUQLTPCYUFPYKE-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C27H25F2N3OS/c1-18-24(26(33)32-16-17-34-27(32)30-18)12-15-31-13-10-21(11-14-31)25(19-2-6-22(28)7-3-19)20-4-8-23(29)9-5-20/h2-9,16-17H,10-15H2,1H3 |
| PubChem CID | 5074 |
| ChEMBL | CHEMBL267777 |
| IUPHAR | 97 |
| BindingDB | 50001775 |
| DrugBank | DB12693 |
Structure | 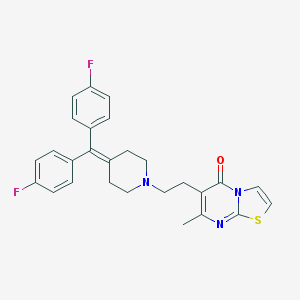 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| IC50 | 4.5 nM | PMID16469866 | BindingDB |
| Ki | 0.25 nM | PMID9225287, PMID7582481 | PDSP,BindingDB |
| Ki | 0.251189 - 6.30957 nM | PMID15322733 | IUPHAR |
| Ki | 0.44 nM | PMID16469866 | BindingDB |
| Ki | 1.62 nM | PMID11882920 | BindingDB |
| Ki | 1.62181 nM | PMID11882920 | PDSP |
| Ki | 1.8 nM | MedChemComm, (2015) 6:4:601, PMID27261181, http://pubs.rsc.org/en/content/articlepdf/2015/md/c4md00418c | PDSP,BindingDB,ChEMBL |
| Ki | 6.60693 nM | PMID15322733 | PDSP |
| Ki | 6.61 nM | PMID15322733 | BindingDB |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417