

You can:
| Name | 5-hydroxytryptamine receptor 2A |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR2A |
| Synonym | 5-HT-2 serotonin receptor 2A serotonin 5HT-2 receptor 5-HT-2A 5-HT2A receptor [ Show all ] |
| Disease | Depression Unspecified Diabetes Erythropoietic porphyria Fibromyalgia [ Show all ] |
| Length | 471 |
| Amino acid sequence | MDILCEENTSLSSTTNSLMQLNDDTRLYSNDFNSGEANTSDAFNWTVDSENRTNLSCEGCLSPSCLSLLHLQEKNWSALLTAVVIILTIAGNILVIMAVSLEKKLQNATNYFLMSLAIADMLLGFLVMPVSMLTILYGYRWPLPSKLCAVWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNSRTKAFLKIIAVWTISVGISMPIPVFGLQDDSKVFKEGSCLLADDNFVLIGSFVSFFIPLTIMVITYFLTIKSLQKEATLCVSDLGTRAKLASFSFLPQSSLSSEKLFQRSIHREPGSYTGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICKESCNEDVIGALLNVFVWIGYLSSAVNPLVYTLFNKTYRSAFSRYIQCQYKENKKPLQLILVNTIPALAYKSSQLQMGQKKNSKQDAKTTDNDCSMVALGKQHSEEASKDNSDGVNEKVSCV |
| UniProt | P28223 |
| Protein Data Bank | 6a93, 6a94 |
| GPCR-HGmod model | P28223 |
| 3D structure model | This structure is from PDB ID 6a93. |
| BioLiP | BL0441025,BL0441028, BL0441031, BL0441030,BL0441033, BL0441029,BL0441032, BL0441026, BL0441024,BL0441027 |
| Therapeutic Target Database | T32060 |
| ChEMBL | CHEMBL224 |
| IUPHAR | 6 |
| DrugBank | BE0000451 |
| Name | ritanserin |
|---|---|
| Molecular formula | C27H25F2N3OS |
| IUPAC name | 6-[2-[4-[bis(4-fluorophenyl)methylidene]piperidin-1-yl]ethyl]-7-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one |
| Molecular weight | 477.574 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 0 |
| XlogP | 5.2 |
| Synonyms | Tiserton CHEMBL267777 SBI-0051053.P003 DivK1c_000192 FT-0630948 [ Show all ] |
| Inchi Key | JUQLTPCYUFPYKE-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C27H25F2N3OS/c1-18-24(26(33)32-16-17-34-27(32)30-18)12-15-31-13-10-21(11-14-31)25(19-2-6-22(28)7-3-19)20-4-8-23(29)9-5-20/h2-9,16-17H,10-15H2,1H3 |
| PubChem CID | 5074 |
| ChEMBL | CHEMBL267777 |
| IUPHAR | 97 |
| BindingDB | 50001775 |
| DrugBank | DB12693 |
Structure | 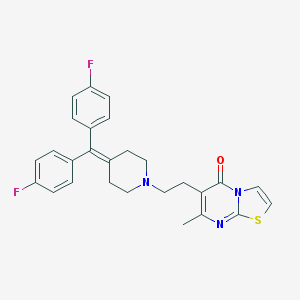 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| IC50 | 0.069 nM | PMID16469866 | BindingDB |
| Ki | 0.01 nM | PMID7582481 | PDSP,BindingDB |
| Ki | 0.083 nM | PMID16469866 | BindingDB |
| Ki | 0.25 nM | PMID9225287 | PDSP,BindingDB |
| Ki | 0.398107 nM | PMID10188965 | IUPHAR |
| Ki | 1.0 nM | PMID7582481 | PDSP,BindingDB |
| Ki | 1.09 nM | PMID7984267 | PDSP,BindingDB |
| Ki | 1.26 nM | Hoyer et al., PMID1987 | PDSP |
| Ki | 4.57 nM | PMID15322733 | BindingDB |
| Ki | 4.57088 nM | PMID15322733 | PDSP |
| Ki | 5.01187 nM | PMID15322733 | IUPHAR |
| Ki | 6.3 nM | Elliot & Kent, PMID1989 | PDSP |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417