
DeepMSA (version 2) is a composite approach to generate high quality multiple sequence alignments for
protein monomers or protein multimers based on huge genomics and metagenomics databases with a structure model-based multi-MSA ranking system.
For protein monomer, the MSAs are produced by three iterative MSA generation pipelines with large alignment depth and diverse sequence sources by merging
sequences from whole-genome sequence databases (Uniclust30 and UniRef90) and from metagenome databases (Metaclust, BFD, Mgnify, TaraDB, MetaSourceDB and JGIclust).
For protein multimer, the top N ranked MSAs for each constituent protein are selected for generating
potential paired MSAs. Each selected MSA for one constituent protein can be paired with the MSA of
another constituent. Large-scale benchmark data show that the performance of several important tasks in protein research, including protein monomer and complex
structure prediction, template detection, and contact/distance prediction, can be significantly improved after utilizing DeepMSA2-derived MSAs.
Methods
DeepMSA (version 2) consists of two separate pipelines for monomer and multimer MSA constructions
respectively. For monomer MSA construction, it utilizes three parallel blocks (dMSA, qMSA, and mMSA)
built on different searching strategies to obtain raw MSAs from a diversity set of databases from
whole-genome and metagenome sequence libraries. In each of the three MSA generation blocks,
a similar logic is followed, in which an initial query is searched against a sequence database,
and if a sufficient number of effective sequences is not achieved, iterative searches into
larger databases are attempted. Finally, up to 10 raw MSAs are scanned and ranked
through a rapid deep learning folding process to select the optimal candidate MSA.
For multimeric MSA construction, multiple composite sequences are created by linking
the monomeric sequences from different component chains that have the same orthologous origins.
Here, a set of M top ranked monomeric MSAs from each chain are paired those of all other chains,
which result in M^m hybrid multimeric MSAs with m being the number of distinct monomer chains
in the complex. The optimal multimer MSAs are then selected based on a combined score of
the depth of the MSAs and folding score of the monomer chains.
Server inputs
The user needs to paste the fasta-formatted amino acid sequence (or sequences for protein complex)
into the input box, or upload the amino acid sequence of the query protein using the "Choose file" button.
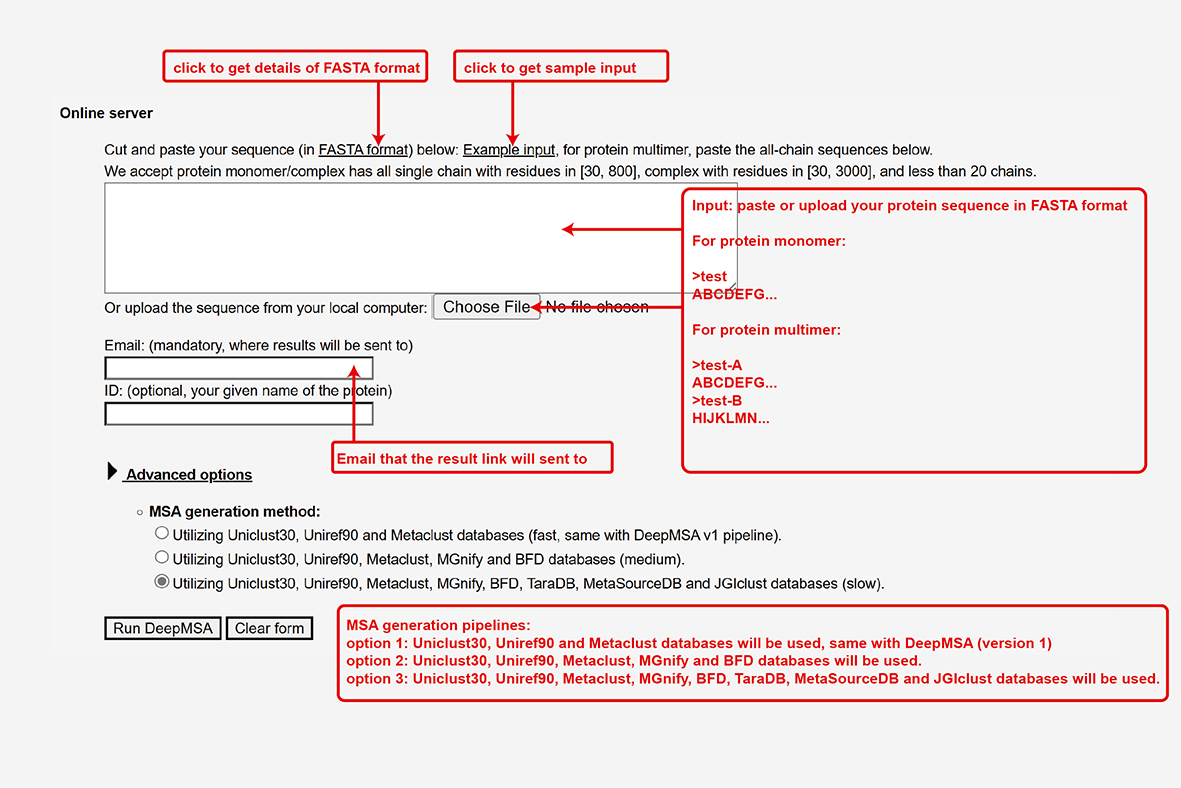
Input of DeepMSA.
Server outputs
For protein monomer:

Output for protein monomer.
For protein multimer:

Output for protein multimer.
How to cite DeepMSA2?
- Wei Zheng, Qiqige Wuyun, Yang Li, Chengxin Zhang, P Lydia Freddolino, Yang Zhang.
Improving deep learning protein monomer and complex structure prediction using DeepMSA2 with huge metagenomics data.
Nature Methods, (2024). https://doi.org/10.1038/s41592-023-02130-4.
[back to server]






![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417