DAMpred for Recognizing Disease-Associated SNP Mutations
DAMpred is a method for predicting Disease-associated mutations of proteins in
the human genome.
A novel training method integrates Bayes classification and artificial neutral
network to comprehensively consider sequence-based, biological assembly-based
and I-TASSER model-based features and their corresponding probability density
on disease-associated dataset and neutral dataset, respectively.
Figure 1 shows the Flowchart of DAMpred for disease-associated mutation prediction.
All the features of DAMpred are derived from protein sequence, so its ability to
deliver good predictions without experimental structure high-quality extend
the application of disease-associated mutation prediction.
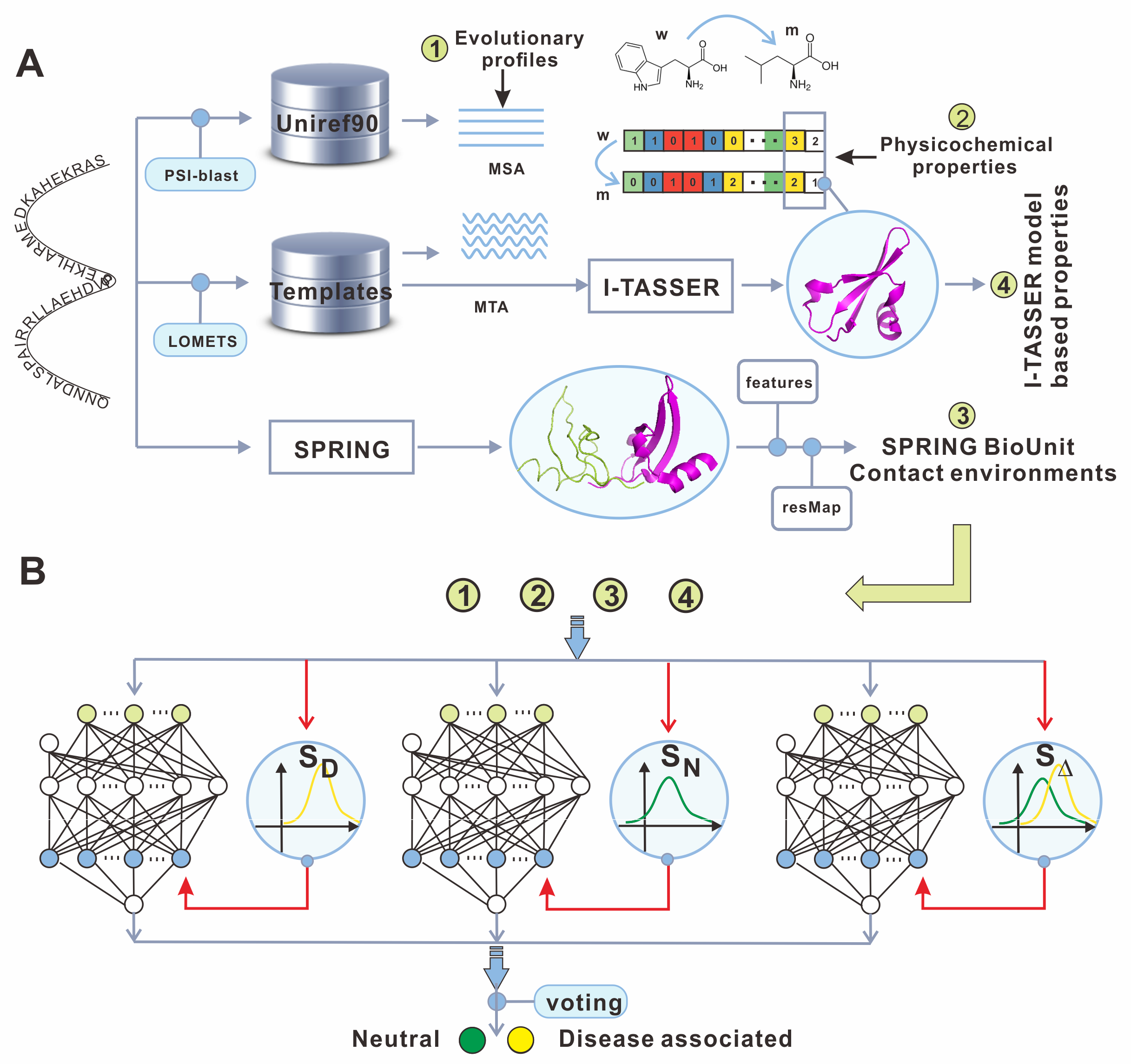 Figure 1: The flowchart of DAMpred method.
Figure 1: The flowchart of DAMpred method.
The DAMpred algorithm was tested on a set of 10,634 mutations from 2,154 proteins.
After excluding homologous templates with sequence identity >30% to the target,
I-TASSER was able to build correct structure model with a TM-score > 0.5 for
1,863 proteins. Using the I-TASSER model, DAMpred classifies all mutation into
two groups, neutral and disease-associated, with an ACC 0.800 and MCC 0.601
in the protein-level 10-fold cross-validation.
DAMpred improve 17% on average MCC, indicating that DAMpred give more balanced accuracy,
especially for neutral mutations, compared with others methods.
How to use Molde: Single-point mutations |
First step: Please copy and paste your data (Note: DAMpred with sequence in FASTA format will run I-TASSER to get 3D structure, which will take more time).
Either sequence in FASTA format is acceptable:
KLHKEPATLIKAIDGDTVKLMYKGQPMTFRLLLVDTPETKHPKKGVEKYGPEASAFTKKM
VENAKKIEVEFDKGQRTDKYGRGLAYIYADGKMVNEALVRQGLAKVAYVYKPNNTHEQHL
RKSEAQAKKEKLNIWS
Second step: Please input a list of mutations
(one per mutation set per line with single-point mutations set out off by semicolons,
WT residue followed by the position of the mutation followed by the mutated residue). Example,
L20A; -Single mutation Lys, the 1th residue, is mutated to Ala
K105M;
Last step: You will get the results as shown (>> Example of Output ...).
|
References:
-
Lijun Quan, Hongjie Qu, Qiang Lyu, Yang Zhang.
DAMpred: Recognizing disease-associated nsSNP mutations through Bayes-guided neural-network model
built on low-resolution structure prediction of proteins and protein-protein interactions,
submitted (2018).
Back to DAMpred



![]() zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417