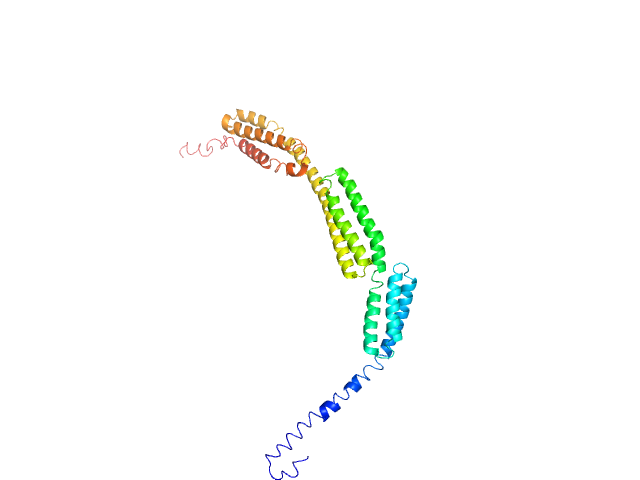| # | Accession, Gene | Matcha,b | Our annotation and structure model | Fmax MF/BP | neXtProt annotation |
| 1 | Q96M27-1, PRRC1 | * | protein kinase A regulation
 | 1.00,0.88 | Activation of protein kinase A activity. Protein binding. Protein kinase A regulatory subunit binding. |
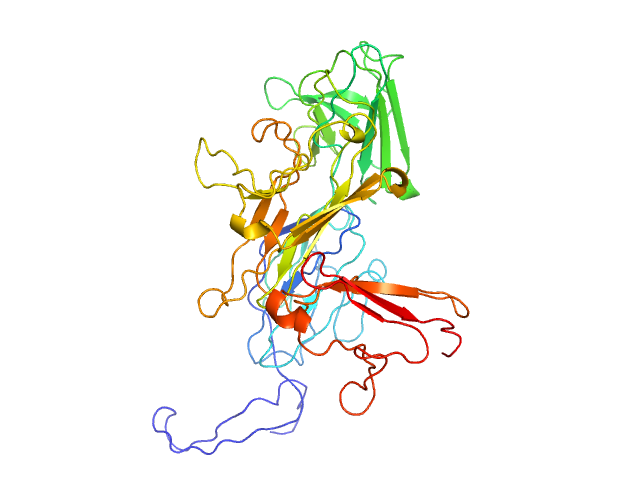
| 2 | P0C870-1, JMJD7 | * | histone demethylation
 | 0.55,0.90 | Bifunctional enzyme that acts both as an endopeptidase and 2-oxoglutarate-dependent monoxygenase. Endopeptidase that cleaves histones N-terminal tails at the carboxyl side of methylated arginine or lysine residues, to generate 'tailless nucleosomes', which may trigger transcription elongation. Preferentially recognizes and cleaves monomethylated and dimethylated arginine residues of histones H2, H3 and H4. After initial cleavage, continues to digest histones tails via its aminopeptidase activity. Additionally, may play a role in protein biosynthesis by modifying the translation machinery. Acts as Fe2+ and 2-oxoglutarate-dependent monoxygenase, catalyzing (S)-stereospecific hydroxylation at C-3 of 'Lys-22' of DRG1 and 'Lys-21' of DRG2 translation factors (TRAFAC), promoting their interaction with ribonucleic acids (RNA). |
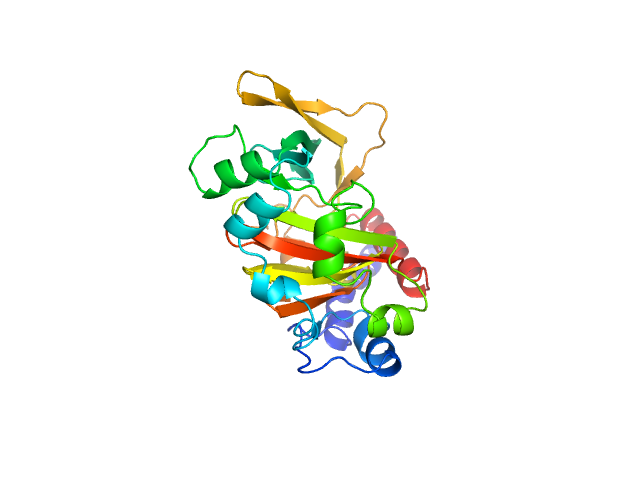
| 3 | Q7Z5A7-1, FAM19A5 | * | regulation of microglial cell activation
 | 0.67,0.50 | Acts as a chemokine-like protein by regulating cell proliferation and migration through activation of G protein-coupled receptors (GPCRs), such as S1PR2 and FPR2. Stimulates chemotactic migration of macrophages mediated by the MAPK3/ERK1 and AKT1 pathway. Blocks TNFSF11/RANKL-induced osteoclast formation from macrophages by inhibiting up-regulation of osteoclast fusogenic and differentiation genes. Stimulation of macrophage migration and inhibition of osteoclast formation is mediated via GPCR FPR2. Acts as an adipokine by negatively regulating vascular smooth muscle cell (VSMC) proliferation and migration in response to platelet-derived growth factor stimulation via GPCR S1PR2 and G protein GNA12/GNA13-transmitted RHOA signaling. Inhibits injury-induced cell proliferation and neointima formation in the femoral arteries |
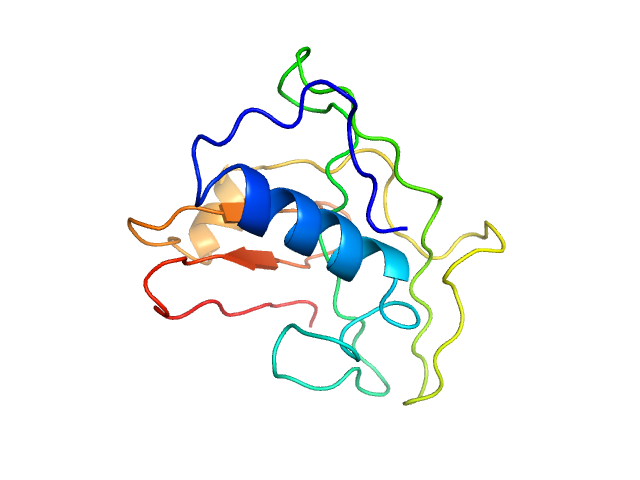
| 4 | Q5T0D9-1, TPRG1L | + | phosphatidylinositol-4-phosphate phosphatase
 | NA,0.55 | Presynaptic protein involved in the synaptic transmission tuning. Regulates synaptic release probability by decreasing the calcium sensitivity of release |
| 5 | Q96D15-1, RCN3 | | catalytic activity, acting on a protein
 | NA,0.47 | Probable molecular chaperone assisting protein biosynthesis and transport in the endoplasmic reticulum. Required for the proper biosynthesis and transport of pulmonary surfactant-associated protein A/SP-A, pulmonary surfactant-associated protein D/SP-D and the lipid transporter ABCA3. By regulating both the proper expression and the degradation through the endoplasmic reticulum-associated protein degradation pathway of these proteins plays a crucial role in pulmonary surfactant homeostasis. Has an anti-fibrotic activity by negatively regulating the secretion of type I and type III collagens. This calcium-binding protein also transiently associates with immature PCSK6 and regulates its secretion. |
| 6 | Q8WTR8-1, NTN5 | * | anatomical structure morphogenesis
 | NA,0.44 | Plays a role in neurogenesis. Prevents motor neuron cell body migration out of the neural tube. |
| 7 | Q9C0D6-1, FHDC1 | * | binding of cytoskeleton
 | 0.44,0.33 | Microtubule-associated formin which regulates both actin and microtubule dynamics. Induces microtubule acetylation and stabilization and actin stress fiber formation. Regulates Golgi ribbon formation. Required for normal cilia assembly. |
| 8 | O75363-1, BCAS1 | | (for CC: neuron part)
 | NA,0.40 | Required for myelination. |
| 9 | P60827-1, C1QTNF8 | * | signaling receptor binding
 | NA,0.40 | May play a role as ligand of relaxin receptor RXFP1. |
| 10 | Q8IUY3-1, GRAMD2A | | binding of GTPase from Ras superfamily
 | 0.11,0.38 | Participates in the organization of endoplasmic reticulum-plasma membrane contact sites (EPCS) with pleiotropic functions including STIM1 recruitment and calcium homeostasis. Constitutive tether that co-localize with ESYT2/3 tethers at endoplasmic reticulum-plasma membrane contact sites in a phosphatidylinositol lipid-dependent manner. Pre-marks the subset of phosphtidylinositol 4,5-biphosphate (PI(4,5)P2)-enriched EPCS destined for the store operated calcium entry pathway (SOCE). |
| 11 | Q9BZH6-1, WDR11 | |
 | NA,0.35 | Involved in the Hedgehog (Hh) signaling pathway, is essential for normal ciliogenesis. Regulates the proteolytic processing of GLI3 and cooperates with the transcription factor EMX1 in the induction of downstream Hh pathway gene expression and gonadotropin-releasing hormone production. WDR11 complex facilitates the tethering of Adaptor protein-1 complex (AP-1)-derived vesicles. WDR11 complex acts together with TBC1D23 to facilitate the golgin-mediated capture of vesicles generated using AP-1. |
| 12 | Q6ZNE9-2, RUFY4 | | regulation of protein folding
 | 0.26,0.32 | Positively regulates macroautophagy in primary dendritic cells. Increases autophagic flux, probably by stimulating both autophagosome formation and facilitating tethering with lysosomes. Binds to phosphatidylinositol 3-phosphate (PtdIns3P) through its FYVE-type zinc finger. |
| 13 | Q9GZU8-1, FAM192A | | hydrolase, probably hydrolase of protein
 | NA,0.32 | Promotes the association of the proteasome activator complex subunit PSME3 with the 20S proteasome and regulates its activity. Inhibits PSME3-mediated degradation of some proteasome substrates, probably by affecting their diffusion rate into the catalytic chamber of the proteasome |
| 14 | Q494U1-1, PLEKHN1 | | transmembrane transport of small molecules, such as nucleotide
 | 0.05,0.29 | Controls the stability of the leptin mRNA harboring an AU-rich element (ARE) in its 3' UTR, in cooperation with the RNA stabilizer ELAVL1 |
| 15 | Q8IUW5-1, RELL1 | * | regulation of apoptosis through TNF
 | NA,0.29 | Induces activation of MAPK14/p38 cascade, when overexpressed. |
| 16 | Q8NDM7-1, CFAP43 | |
 | NA,0.29 | Flagellar protein involved in sperm flagellum axoneme organization and function. |
| 17 | Q8TDG2-1, ACTRT1 | | regulation of chromosome organization either though histone acetylation or binding of cytoskelton used in chromsome segregation
 | 0.03,0.29 | Negatively regulates the Hedgehog (SHH) signaling. Binds to the promoter of the SHH signaling mediator, GLI1, and inhibits its expression. |
| 18 | O75677-1, RFPL1 | * | ubiquitin-protein transferase activity
 | NA,0.27 | Negatively regulates the G2-M phase transition, possibly by promoting cyclin B1/CCNB1 and CDK1 proteasomal degradation and thereby preventing their accumulation during interphase. |
| 19 | Q5VTQ0-1, TTC39B | * | protein ubiquitination regulation
 | NA,0.26 | Regulates high density lipoprotein (HDL) cholesterol metabolism by promoting the ubiquitination and degradation of the oxysterols receptors LXR (NR1H2 and NR1H3). |
| 20 | Q96S16-1, JMJD8 | | histone demethylation
 | NA,0.21 | Functions as a positive regulator of TNF-induced NF-kappa-B signaling. Regulates angiogenesis and cellular metabolism through interaction with PKM. |
| 21 | Q9H9L7-1, AKIRIN1 | | by binding to RNA polymerase, regulate expression of genes such as cytokines
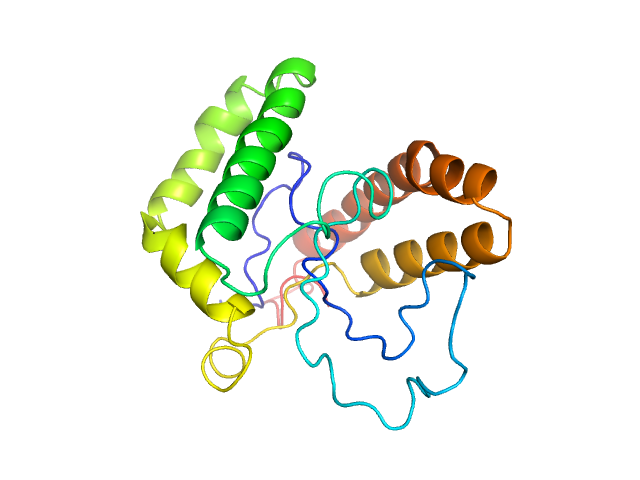 | NA,0.18 | Functions as signal transducer for MSTN during skeletal muscle regeneration and myogenesis. May regulates chemotaxis of both macrophages and myoblasts by reorganising actin cytoskeleton, leading to more efficient lamellipodia formation via a PI3 kinase dependent pathway. |
| 22 | Q96KV7-1, WDR90 | | regulation of transcription by nucleic acid binding
 | NA,0.17 | Required for efficient primary cilium formation. |
| 23 | Q6AI39-1, BICRAL | | sodium:potassium ion transporter
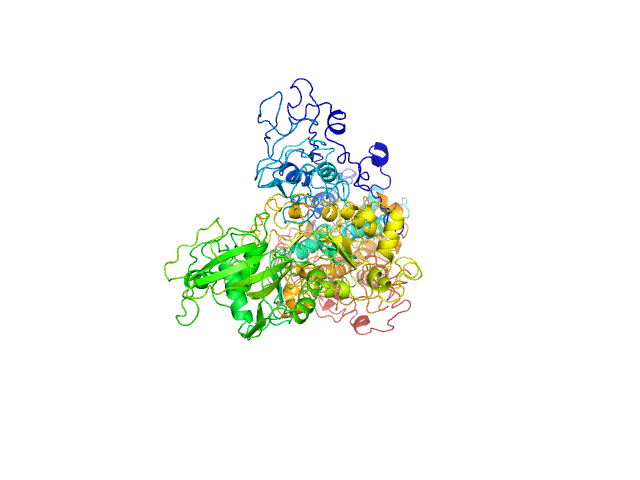 | NA,NA | Component of SWI/SNF chromatin remodeling subcomplex GBAF that carries out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. |
| 24 | Q96J88-1, EPSTI1 | | cytoskeleton binding
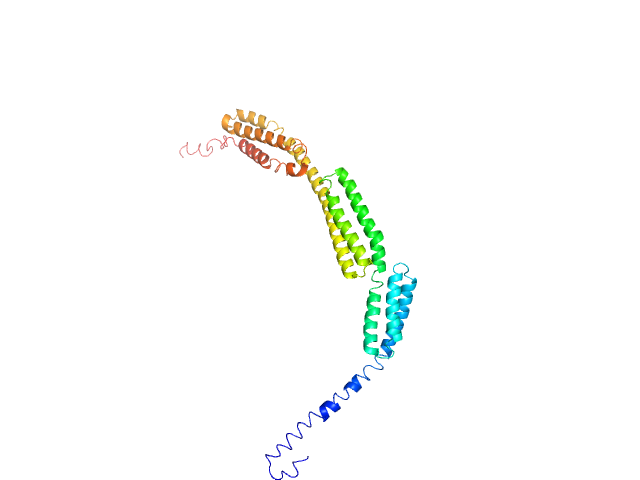 | NA,NA | Plays a role in M1 macrophage polarization and is required for the proper regulation of gene expression during M1 versus M2 macrophage differentiation. Might play a role in RELA/p65 and STAT1 phosphorylation and nuclear localization upon activation of macrophages. |
| 25 | Q9BZD6-1, PRRG4 | | serine-type endopeptidase
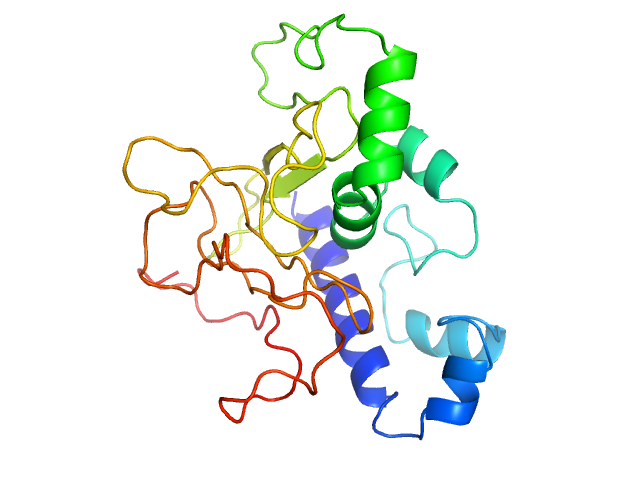 | NA,NA | May control axon guidance across the CNS. Prevents the delivery of ROBO1 at the cell surface and downregulates its expression. |
| # | Accession, Gene (chromosome) | Matcha | Our annotationb | Fmax MF/BPc | neXtProt annotationd |
| 1 |
Q96M27-1, PRRC1 (Chr5) |
* |
protein kinase A regulation
==== MF ====
GO:0034237 F 0.65 protein kinase A regulatory subunit binding

==== BP ====
GO:0065007 P 0.94 biological regulation
GO:0050789 P 0.92 regulation of biological process
GO:0050794 P 0.91 regulation of cellular process
GO:0060255 P 0.82 regulation of macromolecule metabolic process
GO:0031323 P 0.81 regulation of cellular metabolic process
GO:0080090 P 0.80 regulation of primary metabolic process
GO:0051171 P 0.80 regulation of nitrogen compound metabolic process
GO:0048518 P 0.79 positive regulation of biological process
GO:0048522 P 0.77 positive regulation of cellular process
GO:0009987 P 0.77 cellular process
GO:0065008 P 0.74 regulation of biological quality
GO:0032501 P 0.74 multicellular organismal process
GO:0010604 P 0.73 positive regulation of macromolecule metabolic process
GO:0051173 P 0.72 positive regulation of nitrogen compound metabolic process
GO:0031325 P 0.72 positive regulation of cellular metabolic process
GO:0065009 P 0.71 regulation of molecular function
GO:0051246 P 0.71 regulation of protein metabolic process
GO:0032268 P 0.70 regulation of cellular protein metabolic process
GO:0050790 P 0.69 regulation of catalytic activity
GO:0031399 P 0.69 regulation of protein modification process
GO:0051247 P 0.68 positive regulation of protein metabolic process
GO:0044093 P 0.68 positive regulation of molecular function
GO:0019220 P 0.68 regulation of phosphate metabolic process
GO:0045859 P 0.67 regulation of protein kinase activity
GO:0042592 P 0.67 homeostatic process
GO:0001934 P 0.67 positive regulation of protein phosphorylation
GO:0043085 P 0.66 positive regulation of catalytic activity
GO:0007389 P 0.66 pattern specification process
GO:0034199 P 0.65 activation of protein kinase A activity
GO:0010669 P 0.65 epithelial structure maintenance
GO:0001757 P 0.65 somite specification

 |
1.00,0.88 |
Activation of protein kinase A activity. Protein binding. Protein kinase A regulatory subunit binding.
==== MF ====
GO:0034237 ! protein kinase A regulatory subunit binding

==== BP ====
GO:0034199 ! activation of protein kinase A activity
 |
| 2 |
P0C870-1, JMJD7 (Chr15) |
* |
histone demethylation
==== MF ====
GO:0003824 F 0.98 catalytic activity
GO:0016491 F 0.79 oxidoreductase activity
GO:0016705 F 0.78 oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen
GO:0140096 F 0.77 catalytic activity, acting on a protein
GO:0016706 F 0.76 2-oxoglutarate-dependent dioxygenase activity
GO:1901363 F 0.69 heterocyclic compound binding
GO:0097159 F 0.69 organic cyclic compound binding
GO:0032452 F 0.63 histone demethylase activity
GO:0043167 F 0.56 ion binding
GO:0046872 F 0.52 metal ion binding

==== BP ====
GO:0009987 P 1.00 cellular process
GO:0008152 P 0.98 metabolic process
GO:0071704 P 0.97 organic substance metabolic process
GO:0044238 P 0.97 primary metabolic process
GO:0044237 P 0.97 cellular metabolic process
GO:1901564 P 0.96 organonitrogen compound metabolic process
GO:0043412 P 0.95 macromolecule modification
GO:0044260 P 0.84 cellular macromolecule metabolic process
GO:0065007 P 0.78 biological regulation
GO:0050789 P 0.77 regulation of biological process
GO:0006464 P 0.76 cellular protein modification process

 |
0.55,0.90 |
Bifunctional enzyme that acts both as an endopeptidase and 2-oxoglutarate-dependent monoxygenase. Endopeptidase that cleaves histones N-terminal tails at the carboxyl side of methylated arginine or lysine residues, to generate 'tailless nucleosomes', which may trigger transcription elongation. Preferentially recognizes and cleaves monomethylated and dimethylated arginine residues of histones H2, H3 and H4. After initial cleavage, continues to digest histones tails via its aminopeptidase activity. Additionally, may play a role in protein biosynthesis by modifying the translation machinery. Acts as Fe2+ and 2-oxoglutarate-dependent monoxygenase, catalyzing (S)-stereospecific hydroxylation at C-3 of 'Lys-22' of DRG1 and 'Lys-21' of DRG2 translation factors (TRAFAC), promoting their interaction with ribonucleic acids (RNA).
==== MF ====
GO:0016706 ! 2-oxoglutarate-dependent dioxygenase activity
GO:0004177 ! aminopeptidase activity
GO:0004175 ! endopeptidase activity
GO:0046872 ! metal ion binding
GO:0035064 ! methylated histone binding
GO:0004497 ! monooxygenase activity

==== BP ====
GO:0018126 ! protein hydroxylation
 |
| 3 |
Q7Z5A7-1, FAM19A5 (Chr22) |
* |
regulation of microglial cell activation
==== MF ====

==== BP ====
GO:0050789 P 1.00 regulation of biological process
GO:0050794 P 0.85 regulation of cellular process
GO:0009987 P 0.84 cellular process
GO:0048522 P 0.75 positive regulation of cellular process
GO:0048583 P 0.73 regulation of response to stimulus

 |
0.67,0.50 |
Acts as a chemokine-like protein by regulating cell proliferation and migration through activation of G protein-coupled receptors (GPCRs), such as S1PR2 and FPR2. Stimulates chemotactic migration of macrophages mediated by the MAPK3/ERK1 and AKT1 pathway. Blocks TNFSF11/RANKL-induced osteoclast formation from macrophages by inhibiting up-regulation of osteoclast fusogenic and differentiation genes. Stimulation of macrophage migration and inhibition of osteoclast formation is mediated via GPCR FPR2. Acts as an adipokine by negatively regulating vascular smooth muscle cell (VSMC) proliferation and migration in response to platelet-derived growth factor stimulation via GPCR S1PR2 and G protein GNA12/GNA13-transmitted RHOA signaling. Inhibits injury-induced cell proliferation and neointima formation in the femoral arteries
==== MF ====
GO:0005125 ! cytokine activity

==== BP ====
GO:0010469 ! regulation of signaling receptor activity
 |
| 4 |
Q5T0D9-1, TPRG1L (Chr1) |
+ |
phosphatidylinositol-4-phosphate phosphatase
==== BP ====
GO:0065007 P 0.96 biological regulation
GO:0050794 P 0.95 regulation of cellular process

 |
NA,0.55 |
Presynaptic protein involved in the synaptic transmission tuning. Regulates synaptic release probability by decreasing the calcium sensitivity of release
==== BP ====
GO:0051966 ! regulation of synaptic transmission, glutamatergic
 |
| 5 |
Q96D15-1, RCN3 (Chr19) |
|
catalytic activity, acting on a protein
==== BP ====
GO:0009987 P 0.95 cellular process
GO:0065007 P 0.83 biological regulation
GO:0050789 P 0.78 regulation of biological process
GO:0050794 P 0.75 regulation of cellular process
GO:0071704 P 0.66 organic substance metabolic process
GO:0044237 P 0.65 cellular metabolic process
GO:0044238 P 0.64 primary metabolic process
GO:0006807 P 0.63 nitrogen compound metabolic process
GO:0043170 P 0.61 macromolecule metabolic process
GO:0044260 P 0.56 cellular macromolecule metabolic process
GO:1901564 P 0.54 organonitrogen compound metabolic process
GO:0007165 P 0.54 signal transduction
GO:0019538 P 0.52 protein metabolic process
GO:0043412 P 0.51 macromolecule modification

 |
NA,0.47 |
Probable molecular chaperone assisting protein biosynthesis and transport in the endoplasmic reticulum. Required for the proper biosynthesis and transport of pulmonary surfactant-associated protein A/SP-A, pulmonary surfactant-associated protein D/SP-D and the lipid transporter ABCA3. By regulating both the proper expression and the degradation through the endoplasmic reticulum-associated protein degradation pathway of these proteins plays a crucial role in pulmonary surfactant homeostasis. Has an anti-fibrotic activity by negatively regulating the secretion of type I and type III collagens. This calcium-binding protein also transiently associates with immature PCSK6 and regulates its secretion.
==== BP ====
GO:0009306 ! protein secretion
GO:0010952 ! positive regulation of peptidase activity
GO:0015031 ! protein transport
GO:0032964 ! collagen biosynthetic process
GO:0036503 ! ERAD pathway
GO:0043129 ! surfactant homeostasis
GO:0051896 ! regulation of protein kinase B signaling
GO:0055091 ! phospholipid homeostasis
GO:0060428 ! lung epithelium development
 |
| 6 |
Q8WTR8-1, NTN5 (Chr19) |
* |
anatomical structure morphogenesis
==== BP ====
GO:0009987 P 0.98 cellular process
GO:0065007 P 0.88 biological regulation
GO:0050789 P 0.86 regulation of biological process
GO:0032502 P 0.86 developmental process

 |
NA,0.44 |
Plays a role in neurogenesis. Prevents motor neuron cell body migration out of the neural tube.
==== BP ====
GO:0022008 ! neurogenesis
 |
| 7 |
Q9C0D6-1, FHDC1 (Chr4) |
* |
binding of cytoskeleton
==== MF ====
GO:0008092 F 0.70 cytoskeletal protein binding
GO:0003824 F 0.60 catalytic activity

==== BP ====
GO:0009987 P 0.95 cellular process
GO:0065007 P 0.82 biological regulation
GO:0050789 P 0.80 regulation of biological process
GO:0050794 P 0.75 regulation of cellular process
GO:0016043 P 0.69 cellular component organization
GO:0032502 P 0.54 developmental process

 |
0.44,0.33 |
Microtubule-associated formin which regulates both actin and microtubule dynamics. Induces microtubule acetylation and stabilization and actin stress fiber formation. Regulates Golgi ribbon formation. Required for normal cilia assembly.
==== MF ====
GO:0003779 ! actin binding
GO:0008017 ! microtubule binding

==== BP ====
GO:0043149 ! stress fiber assembly
GO:0060271 ! cilium assembly
GO:0090161 ! Golgi ribbon formation
 |
| 8 |
O75363-1, BCAS1 (Chr20) |
|
(for CC: neuron part)
==== BP ====
GO:0009987 P 0.90 cellular process

 |
NA,0.40 |
Required for myelination.
==== BP ====
GO:0042552 ! myelination
 |
| 9 |
P60827-1, C1QTNF8 (Chr16) |
* |
signaling receptor binding
==== BP ====
GO:0009987 P 0.93 cellular process
GO:0065007 P 0.85 biological regulation
GO:0050789 P 0.82 regulation of biological process
GO:0048518 P 0.67 positive regulation of biological process
GO:0032502 P 0.65 developmental process
GO:0050896 P 0.62 response to stimulus
GO:0050794 P 0.62 regulation of cellular process
GO:0048583 P 0.62 regulation of response to stimulus

 |
NA,0.40 |
May play a role as ligand of relaxin receptor RXFP1.
==== BP ====
GO:2000147 ! positive regulation of cell motility
 |
| 10 |
Q8IUY3-1, GRAMD2A (Chr15) |
|
binding of GTPase from Ras superfamily
==== MF ====
GO:0098772 F 0.64 molecular function regulator
GO:0017016 F 0.63 Ras GTPase binding
GO:0005096 F 0.62 GTPase activator activity
GO:0017137 F 0.61 Rab GTPase binding

==== BP ====
GO:0065007 P 0.85 biological regulation
GO:0051179 P 0.84 localization
GO:0051641 P 0.83 cellular localization
GO:0050794 P 0.82 regulation of cellular process

 |
0.11,0.38 |
Participates in the organization of endoplasmic reticulum-plasma membrane contact sites (EPCS) with pleiotropic functions including STIM1 recruitment and calcium homeostasis. Constitutive tether that co-localize with ESYT2/3 tethers at endoplasmic reticulum-plasma membrane contact sites in a phosphatidylinositol lipid-dependent manner. Pre-marks the subset of phosphtidylinositol 4,5-biphosphate (PI(4,5)P2)-enriched EPCS destined for the store operated calcium entry pathway (SOCE).
==== MF ====
GO:0005546 ! phosphatidylinositol-4,5-bisphosphate binding
GO:0035091 ! phosphatidylinositol binding

==== BP ====
GO:0061817 ! endoplasmic reticulum-plasma membrane tethering
GO:2001256 ! regulation of store-operated calcium entry
 |
| 11 |
Q9BZH6-1, WDR11 (Chr10) |
|
==== BP ====
GO:0009987 P 0.97 cellular process
GO:0065007 P 0.85 biological regulation
GO:0050789 P 0.82 regulation of biological process
GO:0050794 P 0.80 regulation of cellular process
GO:0044237 P 0.75 cellular metabolic process
GO:0071704 P 0.73 organic substance metabolic process
GO:0044238 P 0.73 primary metabolic process
GO:0071840 P 0.72 cellular component organization or biogenesis
GO:0006807 P 0.72 nitrogen compound metabolic process
GO:0043170 P 0.71 macromolecule metabolic process
GO:0016043 P 0.69 cellular component organization
GO:0048518 P 0.63 positive regulation of biological process
GO:0019222 P 0.62 regulation of metabolic process
GO:0060255 P 0.61 regulation of macromolecule metabolic process
GO:0031323 P 0.61 regulation of cellular metabolic process
GO:0080090 P 0.60 regulation of primary metabolic process
GO:0051171 P 0.60 regulation of nitrogen compound metabolic process
GO:0048522 P 0.59 positive regulation of cellular process
GO:0050896 P 0.58 response to stimulus
GO:1901360 P 0.56 organic cyclic compound metabolic process
GO:0044260 P 0.56 cellular macromolecule metabolic process
GO:0034641 P 0.56 cellular nitrogen compound metabolic process
GO:0032502 P 0.56 developmental process
GO:0006725 P 0.56 cellular aromatic compound metabolic process
GO:0046483 P 0.55 heterocycle metabolic process
GO:0022607 P 0.55 cellular component assembly
GO:0090304 P 0.54 nucleic acid metabolic process
GO:0007165 P 0.54 signal transduction
GO:0019219 P 0.53 regulation of nucleobase-containing compound metabolic process
GO:0048583 P 0.51 regulation of response to stimulus
GO:0009893 P 0.51 positive regulation of metabolic process
GO:0051173 P 0.50 positive regulation of nitrogen compound metabolic process
GO:0043933 P 0.50 protein-containing complex subunit organization
GO:0031325 P 0.50 positive regulation of cellular metabolic process
GO:0010604 P 0.50 positive regulation of macromolecule metabolic process

 |
NA,0.35 |
Involved in the Hedgehog (Hh) signaling pathway, is essential for normal ciliogenesis. Regulates the proteolytic processing of GLI3 and cooperates with the transcription factor EMX1 in the induction of downstream Hh pathway gene expression and gonadotropin-releasing hormone production. WDR11 complex facilitates the tethering of Adaptor protein-1 complex (AP-1)-derived vesicles. WDR11 complex acts together with TBC1D23 to facilitate the golgin-mediated capture of vesicles generated using AP-1.
==== BP ====
GO:0006886 ! intracellular protein transport
GO:0007507 ! heart development
GO:0008589 ! regulation of smoothened signaling pathway
GO:0035264 ! multicellular organism growth
GO:0060271 ! cilium assembly
GO:0060322 ! head development
GO:0099041 ! vesicle tethering to Golgi
 |
| 12 |
Q6ZNE9-2, RUFY4 (Chr2) |
|
regulation of protein folding
==== MF ====
GO:0003824 F 0.71 catalytic activity

==== BP ====
GO:0009987 P 0.96 cellular process
GO:0065007 P 0.78 biological regulation
GO:0071704 P 0.76 organic substance metabolic process
GO:0044238 P 0.76 primary metabolic process
GO:0044237 P 0.76 cellular metabolic process
GO:0050794 P 0.75 regulation of cellular process
GO:0006807 P 0.75 nitrogen compound metabolic process
GO:0006139 P 0.74 nucleobase-containing compound metabolic process
GO:0034654 P 0.71 nucleobase-containing compound biosynthetic process
GO:0051179 P 0.69 localization
GO:0051128 P 0.68 regulation of cellular component organization
GO:0006810 P 0.68 transport
GO:0055086 P 0.63 nucleobase-containing small molecule metabolic process
GO:0051641 P 0.63 cellular localization
GO:0006793 P 0.63 phosphorus metabolic process
GO:0046907 P 0.57 intracellular transport
GO:0031323 P 0.54 regulation of cellular metabolic process
GO:1901135 P 0.53 carbohydrate derivative metabolic process
GO:0061077 P 0.51 chaperone-mediated protein folding
GO:0006458 P 0.51 'de novo' protein folding

 |
0.26,0.32 |
Positively regulates macroautophagy in primary dendritic cells. Increases autophagic flux, probably by stimulating both autophagosome formation and facilitating tethering with lysosomes. Binds to phosphatidylinositol 3-phosphate (PtdIns3P) through its FYVE-type zinc finger.
==== MF ====
GO:0032266 ! phosphatidylinositol-3-phosphate binding

==== BP ====
GO:0000045 ! autophagosome assembly
GO:0016239 ! positive regulation of macroautophagy
GO:0071353 ! cellular response to interleukin-4
 |
| 13 |
Q9GZU8-1, FAM192A (Chr16) |
|
hydrolase, probably hydrolase of protein
==== BP ====
GO:0009987 P 0.88 cellular process
GO:0065007 P 0.82 biological regulation
GO:0050789 P 0.80 regulation of biological process
GO:0050794 P 0.79 regulation of cellular process
GO:0008152 P 0.70 metabolic process
GO:0071704 P 0.67 organic substance metabolic process
GO:0044238 P 0.67 primary metabolic process
GO:0006807 P 0.65 nitrogen compound metabolic process
GO:0043170 P 0.61 macromolecule metabolic process
GO:1901564 P 0.55 organonitrogen compound metabolic process
GO:0071840 P 0.53 cellular component organization or biogenesis
GO:0048519 P 0.52 negative regulation of biological process
GO:0016043 P 0.52 cellular component organization
GO:0048523 P 0.51 negative regulation of cellular process
GO:0044237 P 0.51 cellular metabolic process
GO:0019538 P 0.50 protein metabolic process

 |
NA,0.32 |
Promotes the association of the proteasome activator complex subunit PSME3 with the 20S proteasome and regulates its activity. Inhibits PSME3-mediated degradation of some proteasome substrates, probably by affecting their diffusion rate into the catalytic chamber of the proteasome
==== BP ====
GO:0032091 ! negative regulation of protein binding
GO:1901799 ! negative regulation of proteasomal protein catabolic process
 |
| 14 |
Q494U1-1, PLEKHN1 (Chr1) |
|
transmembrane transport of small molecules, such as nucleotide
==== MF ====
GO:0022857 F 0.93 transmembrane transporter activity
GO:0015075 F 0.76 ion transmembrane transporter activity
GO:0008509 F 0.75 anion transmembrane transporter activity
GO:0008514 F 0.72 organic anion transmembrane transporter activity
GO:0015605 F 0.63 organophosphate ester transmembrane transporter activity
GO:0015318 F 0.63 inorganic molecular entity transmembrane transporter activity
GO:0008028 F 0.59 monocarboxylic acid transmembrane transporter activity

==== BP ====
GO:0006810 P 0.96 transport
GO:0071702 P 0.93 organic substance transport
GO:0055085 P 0.87 transmembrane transport
GO:0009987 P 0.86 cellular process
GO:0006811 P 0.78 ion transport
GO:0034220 P 0.77 ion transmembrane transport
GO:0015711 P 0.77 organic anion transport
GO:0098656 P 0.76 anion transmembrane transport
GO:0071705 P 0.71 nitrogen compound transport
GO:1901264 P 0.68 carbohydrate derivative transport
GO:0065007 P 0.62 biological regulation
GO:0051641 P 0.57 cellular localization
GO:0050789 P 0.57 regulation of biological process
GO:0050794 P 0.56 regulation of cellular process
GO:0046907 P 0.56 intracellular transport
GO:0015931 P 0.55 nucleobase-containing compound transport
GO:0007017 P 0.55 microtubule-based process
GO:0006928 P 0.55 movement of cell or subcellular component
GO:0046903 P 0.53 secretion
GO:0030705 P 0.53 cytoskeleton-dependent intracellular transport
GO:0015780 P 0.53 nucleotide-sugar transmembrane transport

 |
0.05,0.29 |
Controls the stability of the leptin mRNA harboring an AU-rich element (ARE) in its 3' UTR, in cooperation with the RNA stabilizer ELAVL1
==== MF ====
GO:0001786 ! phosphatidylserine binding
GO:0070300 ! phosphatidic acid binding
GO:1901612 ! cardiolipin binding
GO:1901981 ! phosphatidylinositol phosphate binding

==== BP ====
GO:0001666 ! response to hypoxia
GO:0043065 ! positive regulation of apoptotic process
GO:0061158 ! 3'-UTR-mediated mRNA destabilization
 |
| 15 |
Q8IUW5-1, RELL1 (Chr4) |
* |
regulation of apoptosis through TNF
==== BP ====
GO:0050789 P 1.00 regulation of biological process
GO:0009987 P 0.70 cellular process
GO:0050794 P 0.65 regulation of cellular process
GO:0048518 P 0.64 positive regulation of biological process
GO:0097190 P 0.51 apoptotic signaling pathway
GO:0042981 P 0.51 regulation of apoptotic process
GO:0042127 P 0.51 regulation of cell proliferation
GO:0032496 P 0.51 response to lipopolysaccharide
GO:0007275 P 0.51 multicellular organism development
GO:0006955 P 0.51 immune response
GO:0006954 P 0.51 inflammatory response

 |
NA,0.29 |
Induces activation of MAPK14/p38 cascade, when overexpressed
==== BP ====
GO:1900745 ! positive regulation of p38MAPK cascade
 |
| 16 |
Q8NDM7-1, CFAP43 (Chr10) |
|
==== BP ====
GO:0009987 P 0.98 cellular process
GO:0065007 P 0.70 biological regulation
GO:0050789 P 0.68 regulation of biological process
GO:0050794 P 0.66 regulation of cellular process
GO:0071840 P 0.61 cellular component organization or biogenesis
GO:0016043 P 0.60 cellular component organization

 |
NA,0.29 |
Flagellar protein involved in sperm flagellum axoneme organization and function.
==== BP ====
GO:0007288 ! sperm axoneme assembly
 |
| 17 |
Q8TDG2-1, ACTRT1 (ChrX) |
|
regulation of chromosome organization either though histone acetylation or binding of cytoskelton used in chromsome segregation
==== MF ====
GO:0005198 F 0.90 structural molecule activity
GO:1901363 F 0.89 heterocyclic compound binding
GO:0097159 F 0.89 organic cyclic compound binding
GO:0005200 F 0.89 structural constituent of cytoskeleton
GO:0043167 F 0.85 ion binding
GO:0035639 F 0.84 purine ribonucleoside triphosphate binding
GO:0032555 F 0.84 purine ribonucleotide binding
GO:0005524 F 0.83 ATP binding
GO:0003824 F 0.77 catalytic activity
GO:0016740 F 0.66 transferase activity
GO:0042802 F 0.64 identical protein binding
GO:0004402 F 0.61 histone acetyltransferase activity
GO:0008092 F 0.59 cytoskeletal protein binding
GO:0003676 F 0.51 nucleic acid binding

==== BP ====
GO:0065007 P 0.96 biological regulation
GO:0050789 P 0.95 regulation of biological process
GO:0009987 P 0.90 cellular process
GO:0060255 P 0.89 regulation of macromolecule metabolic process
GO:0048518 P 0.89 positive regulation of biological process
GO:0010468 P 0.88 regulation of gene expression
GO:0009893 P 0.85 positive regulation of metabolic process
GO:0016043 P 0.78 cellular component organization
GO:0010604 P 0.78 positive regulation of macromolecule metabolic process
GO:0010628 P 0.77 positive regulation of gene expression
GO:0032501 P 0.76 multicellular organismal process
GO:0050794 P 0.74 regulation of cellular process
GO:0090131 P 0.71 mesenchyme migration
GO:0032502 P 0.71 developmental process
GO:0044237 P 0.70 cellular metabolic process
GO:0071704 P 0.69 organic substance metabolic process
GO:0044238 P 0.69 primary metabolic process
GO:0006996 P 0.69 organelle organization
GO:0050896 P 0.68 response to stimulus
GO:0006807 P 0.68 nitrogen compound metabolic process
GO:1901564 P 0.67 organonitrogen compound metabolic process
GO:0044260 P 0.67 cellular macromolecule metabolic process
GO:0022607 P 0.66 cellular component assembly
GO:0051716 P 0.65 cellular response to stimulus
GO:0048869 P 0.65 cellular developmental process
GO:0044267 P 0.65 cellular protein metabolic process
GO:0043412 P 0.65 macromolecule modification
GO:0042221 P 0.64 response to chemical
GO:0006950 P 0.64 response to stress
GO:0006464 P 0.64 cellular protein modification process
GO:0006325 P 0.64 chromatin organization
GO:1901360 P 0.63 organic cyclic compound metabolic process
GO:0034641 P 0.63 cellular nitrogen compound metabolic process
GO:0022402 P 0.63 cell cycle process
GO:0070887 P 0.62 cellular response to chemical stimulus
GO:0033554 P 0.62 cellular response to stress
GO:0022414 P 0.62 reproductive process
GO:0006139 P 0.62 nucleobase-containing compound metabolic process
GO:0090304 P 0.61 nucleic acid metabolic process
GO:0048646 P 0.61 anatomical structure formation involved in morphogenesis
GO:0018193 P 0.61 peptidyl-amino acid modification
GO:0016570 P 0.61 histone modification
GO:0003006 P 0.61 developmental process involved in reproduction
GO:1903046 P 0.60 meiotic cell cycle process
GO:0032506 P 0.60 cytokinetic process
GO:0016573 P 0.60 histone acetylation
GO:0010927 P 0.60 cellular component assembly involved in morphogenesis
GO:0006281 P 0.60 DNA repair
GO:0034599 P 0.59 cellular response to oxidative stress
GO:0030476 P 0.59 ascospore wall assembly
GO:0001300 P 0.59 chronological cell aging
GO:0000916 P 0.59 actomyosin contractile ring contraction
GO:0000011 P 0.59 vacuole inheritance
GO:0080090 P 0.57 regulation of primary metabolic process
GO:0051171 P 0.57 regulation of nitrogen compound metabolic process
GO:0031323 P 0.57 regulation of cellular metabolic process
GO:0031326 P 0.54 regulation of cellular biosynthetic process
GO:0019219 P 0.54 regulation of nucleobase-containing compound metabolic process
GO:0010556 P 0.54 regulation of macromolecule biosynthetic process
GO:0048522 P 0.53 positive regulation of cellular process
GO:0048519 P 0.53 negative regulation of biological process
GO:0006355 P 0.53 regulation of transcription, DNA-templated
GO:0051179 P 0.51 localization
GO:0048523 P 0.51 negative regulation of cellular process

 |
0.03,0.29 |
Negatively regulates the Hedgehog (SHH) signaling. Binds to the promoter of the SHH signaling mediator, GLI1, and inhibits its expression.
==== MF ====
GO:0003682 ! chromatin binding

==== BP ====
GO:0008589 ! regulation of smoothened signaling pathway
GO:0045892 ! negative regulation of transcription, DNA-templated
 |
| 18 |
O75677-1, RFPL1 (Chr22) |
* |
ubiquitin-protein transferase activity
==== BP ====
GO:0009987 P 0.97 cellular process
GO:0065007 P 0.93 biological regulation
GO:0050789 P 0.92 regulation of biological process
GO:0050794 P 0.88 regulation of cellular process
GO:0008152 P 0.83 metabolic process
GO:0044237 P 0.82 cellular metabolic process
GO:0071704 P 0.75 organic substance metabolic process
GO:0044238 P 0.74 primary metabolic process
GO:0006807 P 0.72 nitrogen compound metabolic process
GO:0048583 P 0.71 regulation of response to stimulus
GO:0043170 P 0.70 macromolecule metabolic process
GO:0048519 P 0.69 negative regulation of biological process
GO:0048518 P 0.69 positive regulation of biological process
GO:0019222 P 0.69 regulation of metabolic process
GO:0050896 P 0.68 response to stimulus
GO:0016043 P 0.68 cellular component organization
GO:1901564 P 0.67 organonitrogen compound metabolic process
GO:0031323 P 0.67 regulation of cellular metabolic process
GO:0060255 P 0.66 regulation of macromolecule metabolic process
GO:0044260 P 0.66 cellular macromolecule metabolic process
GO:0080090 P 0.65 regulation of primary metabolic process
GO:0051171 P 0.65 regulation of nitrogen compound metabolic process
GO:0023051 P 0.65 regulation of signaling
GO:0019538 P 0.65 protein metabolic process
GO:0010646 P 0.65 regulation of cell communication
GO:0009966 P 0.64 regulation of signal transduction
GO:0002376 P 0.63 immune system process
GO:0048523 P 0.61 negative regulation of cellular process
GO:0006950 P 0.61 response to stress
GO:0048522 P 0.59 positive regulation of cellular process
GO:0044267 P 0.59 cellular protein metabolic process
GO:0043412 P 0.59 macromolecule modification
GO:0006464 P 0.58 cellular protein modification process
GO:0010468 P 0.56 regulation of gene expression
GO:0016567 P 0.55 protein ubiquitination
GO:0051252 P 0.54 regulation of RNA metabolic process
GO:0031326 P 0.54 regulation of cellular biosynthetic process
GO:0006355 P 0.53 regulation of transcription, DNA-templated
GO:0006952 P 0.51 defense response

 |
NA,0.27 |
Negatively regulates the G2-M phase transition, possibly by promoting cyclin B1/CCNB1 and CDK1 proteasomal degradation and thereby preventing their accumulation during interphase.
==== BP ====
GO:0007049 ! cell cycle
GO:0008285 ! negative regulation of cell proliferation
GO:0010972 ! negative regulation of G2/M transition of mitotic cell cycle
GO:0032436 ! positive regulation of proteasomal ubiquitin-dependent protein catabolic process
GO:0043065 ! positive regulation of apoptotic process
GO:0045930 ! negative regulation of mitotic cell cycle
GO:0051782 ! negative regulation of cell division
GO:2001272 ! positive regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis
 |
| 19 |
Q5VTQ0-1, TTC39B (Chr9) |
* |
protein ubiquitination regulation
==== BP ====
GO:0009987 P 0.96 cellular process
GO:0065007 P 0.81 biological regulation
GO:0050789 P 0.79 regulation of biological process
GO:0050794 P 0.76 regulation of cellular process
GO:0008152 P 0.73 metabolic process
GO:0071704 P 0.72 organic substance metabolic process
GO:0044237 P 0.72 cellular metabolic process
GO:0044238 P 0.71 primary metabolic process

 |
NA,0.26 |
Regulates high density lipoprotein (HDL) cholesterol metabolism by promoting the ubiquitination and degradation of the oxysterols receptors LXR (NR1H2 and NR1H3).
==== BP ====
GO:0006629 ! lipid metabolic process
GO:0010874 ! regulation of cholesterol efflux
GO:0010887 ! negative regulation of cholesterol storage
GO:0042632 ! cholesterol homeostasis
GO:0090181 ! regulation of cholesterol metabolic process
 |
| 20 |
Q96S16-1, JMJD8 (Chr16) |
|
histone demethylation
==== BP ====
GO:0009987 P 0.99 cellular process
GO:0071704 P 0.97 organic substance metabolic process
GO:0044238 P 0.97 primary metabolic process
GO:0044237 P 0.97 cellular metabolic process
GO:1901564 P 0.96 organonitrogen compound metabolic process
GO:0043170 P 0.96 macromolecule metabolic process
GO:0043412 P 0.95 macromolecule modification
GO:0044260 P 0.92 cellular macromolecule metabolic process
GO:0019538 P 0.89 protein metabolic process
GO:0006464 P 0.88 cellular protein modification process
GO:0050789 P 0.84 regulation of biological process
GO:0050794 P 0.77 regulation of cellular process
GO:0019222 P 0.76 regulation of metabolic process
GO:0080090 P 0.75 regulation of primary metabolic process
GO:0060255 P 0.75 regulation of macromolecule metabolic process
GO:0031323 P 0.75 regulation of cellular metabolic process
GO:0051171 P 0.74 regulation of nitrogen compound metabolic process
GO:0019219 P 0.71 regulation of nucleobase-containing compound metabolic process
GO:0010468 P 0.71 regulation of gene expression
GO:0051252 P 0.70 regulation of RNA metabolic process
GO:0016043 P 0.67 cellular component organization
GO:0018193 P 0.63 peptidyl-amino acid modification
GO:0006325 P 0.63 chromatin organization
GO:0016570 P 0.61 histone modification
GO:0016577 P 0.52 histone demethylation
GO:0031326 P 0.50 regulation of cellular biosynthetic process

 |
NA,0.21 |
Functions as a positive regulator of TNF-induced NF-kappa-B signaling. Regulates angiogenesis and cellular metabolism through interaction with PKM.
==== BP ====
GO:0006110 ! regulation of glycolytic process
GO:0043123 ! positive regulation of I-kappaB kinase/NF-kappaB signaling
GO:1903302 ! regulation of pyruvate kinase activity
GO:1903672 ! positive regulation of sprouting angiogenesis
 |
| 21 |
Q9H9L7-1, AKIRIN1 (Chr1) |
|
by binding to RNA polymerase, regulate expression of genes such as cytokines
==== BP ====
GO:0045944 P 1.00 positive regulation of transcription by RNA polymerase II
GO:0050896 P 0.91 response to stimulus
GO:0009605 P 0.90 response to external stimulus
GO:0043207 P 0.89 response to external biotic stimulus
GO:0009987 P 0.85 cellular process
GO:1901700 P 0.71 response to oxygen-containing compound
GO:0051239 P 0.71 regulation of multicellular organismal process
GO:0010033 P 0.71 response to organic substance
GO:0051240 P 0.70 positive regulation of multicellular organismal process
GO:0033993 P 0.69 response to lipid
GO:0032496 P 0.68 response to lipopolysaccharide
GO:0001819 P 0.68 positive regulation of cytokine production
GO:0032755 P 0.67 positive regulation of interleukin-6 production
GO:0032502 P 0.65 developmental process
GO:0048856 P 0.61 anatomical structure development

 |
NA,0.18 |
Functions as signal transducer for MSTN during skeletal muscle regeneration and myogenesis. May regulates chemotaxis of both macrophages and myoblasts by reorganising actin cytoskeleton, leading to more efficient lamellipodia formation via a PI3 kinase dependent pathway.
==== BP ====
GO:0010592 ! positive regulation of lamellipodium assembly
GO:0010759 ! positive regulation of macrophage chemotaxis
GO:0014839 ! myoblast migration involved in skeletal muscle regeneration
GO:0045663 ! positive regulation of myoblast differentiation
GO:1902723 ! negative regulation of skeletal muscle satellite cell proliferation
GO:1902725 ! negative regulation of satellite cell differentiation
 |
| 22 |
Q96KV7-1, WDR90 (Chr16) |
|
regulation of transcription by nucleic acid binding
==== BP ====
GO:0009987 P 0.92 cellular process
GO:0065007 P 0.84 biological regulation
GO:0050789 P 0.82 regulation of biological process
GO:0050794 P 0.80 regulation of cellular process
GO:0019222 P 0.66 regulation of metabolic process
GO:0060255 P 0.64 regulation of macromolecule metabolic process
GO:0080090 P 0.63 regulation of primary metabolic process
GO:0031323 P 0.63 regulation of cellular metabolic process
GO:0051171 P 0.62 regulation of nitrogen compound metabolic process
GO:0010468 P 0.60 regulation of gene expression
GO:0019219 P 0.59 regulation of nucleobase-containing compound metabolic process
GO:2000112 P 0.58 regulation of cellular macromolecule biosynthetic process
GO:0051252 P 0.58 regulation of RNA metabolic process
GO:0071840 P 0.57 cellular component organization or biogenesis
GO:0006355 P 0.57 regulation of transcription, DNA-templated
GO:0044237 P 0.56 cellular metabolic process
GO:0016043 P 0.54 cellular component organization

 |
NA,0.17 |
Required for efficient primary cilium formation.
==== BP ====
GO:0060271 ! cilium assembly
 |
| 23 |
Q6AI39-1, BICRAL (Chr6) |
|
sodium:potassium ion transporter
 |
NA,NA |
Component of SWI/SNF chromatin remodeling subcomplex GBAF that carries out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. |
| 24 |
Q96J88-1, EPSTI1 (Chr13) |
|
cytoskeleton binding
 |
NA,NA |
Plays a role in M1 macrophage polarization and is required for the proper regulation of gene expression during M1 versus M2 macrophage differentiation. Might play a role in RELA/p65 and STAT1 phosphorylation and nuclear localization upon activation of macrophages. |
| 25 |
Q9BZD6-1, PRRG4 (Chr11) |
|
serine-type endopeptidase
 |
NA,NA |
May control axon guidance across the CNS. Prevents the delivery of ROBO1 at the cell surface and downregulates its expression. |
Table 3. Predicted GO terms by I-TASSER/COFACTOR for all 44 neXtProt proteins undergoing literature curation as of 2018.
The targets are ranked in alphabetical order of the accession and separated by four-dollar ($$) signs.
Information identical to this table is also provided in a zip file to facilitate programmatic parsing
of the findings (GOterm.zip).
A6NI56-1 transferase acting on a protein
==== MF ====
* GO:0140096 0.50 catalytic activity, acting on a protein
* GO:0016740 0.48 transferase activity
==== BP ====
GO:0009987 0.98 cellular process
GO:0065007 0.81 biological regulation
GO:0050789 0.78 regulation of biological process
GO:0050794 0.77 regulation of cellular process
GO:0051179 0.70 localization
GO:0071840 0.68 cellular component organization or biogenesis
GO:0016043 0.67 cellular component organization
GO:0008152 0.63 metabolic process
GO:0044237 0.61 cellular metabolic process
GO:0048518 0.59 positive regulation of biological process
GO:0071704 0.58 organic substance metabolic process
GO:0048522 0.58 positive regulation of cellular process
GO:0044238 0.57 primary metabolic process
GO:0051128 0.56 regulation of cellular component organization
GO:0043170 0.56 macromolecule metabolic process
GO:0006807 0.56 nitrogen compound metabolic process
GO:1901564 0.52 organonitrogen compound metabolic process
GO:0044260 0.51 cellular macromolecule metabolic process
GO:0022402 0.51 cell cycle process
GO:0019538 0.51 protein metabolic process
==== CC ====
GO:0044424 1.00 intracellular part
GO:0043226 0.89 organelle
GO:0044446 0.87 intracellular organelle part
GO:0044444 0.87 cytoplasmic part
GO:0043227 0.83 membrane-bounded organelle
GO:0043229 0.81 intracellular organelle
GO:0032991 0.75 protein-containing complex
GO:0043231 0.71 intracellular membrane-bounded organelle
GO:0005737 0.52 cytoplasm
GO:0044430 0.51 cytoskeletal part
$$$$
A6NLU0-1 ubiquitin-protein transferase
==== MF ====
GO:0016740 0.83 transferase activity
GO:0140096 0.82 catalytic activity, acting on a protein
* GO:0004842 0.80 ubiquitin-protein transferase activity
==== BP ====
GO:0009987 0.96 cellular process
GO:0050789 0.94 regulation of biological process
GO:0050794 0.90 regulation of cellular process
GO:0008152 0.80 metabolic process
GO:0044237 0.78 cellular metabolic process
GO:0048583 0.76 regulation of response to stimulus
GO:0048519 0.75 negative regulation of biological process
GO:0071704 0.72 organic substance metabolic process
GO:0050896 0.71 response to stimulus
GO:0044238 0.71 primary metabolic process
GO:0023051 0.71 regulation of signaling
GO:0016043 0.71 cellular component organization
GO:0010646 0.71 regulation of cell communication
GO:0019222 0.70 regulation of metabolic process
GO:0009966 0.70 regulation of signal transduction
GO:0048518 0.69 positive regulation of biological process
GO:0006807 0.69 nitrogen compound metabolic process
GO:0048523 0.68 negative regulation of cellular process
GO:0043170 0.68 macromolecule metabolic process
GO:0031323 0.67 regulation of cellular metabolic process
GO:0080090 0.65 regulation of primary metabolic process
GO:0060255 0.65 regulation of macromolecule metabolic process
GO:0044260 0.65 cellular macromolecule metabolic process
GO:0006950 0.65 response to stress
GO:0051171 0.64 regulation of nitrogen compound metabolic process
GO:1901564 0.63 organonitrogen compound metabolic process
GO:0019538 0.61 protein metabolic process
GO:0048522 0.59 positive regulation of cellular process
GO:0043412 0.57 macromolecule modification
GO:0006464 0.56 cellular protein modification process
GO:0010468 0.55 regulation of gene expression
GO:0002376 0.55 immune system process
GO:0019219 0.54 regulation of nucleobase-containing compound metabolic process
GO:2000112 0.53 regulation of cellular macromolecule biosynthetic process
GO:0051252 0.53 regulation of RNA metabolic process
GO:0048585 0.52 negative regulation of response to stimulus
GO:0007166 0.52 cell surface receptor signaling pathway
* GO:0016567 0.51 protein ubiquitination
GO:0006355 0.51 regulation of transcription, DNA-templated
GO:0009968 0.50 negative regulation of signal transduction
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.99 intracellular part
GO:0043226 0.83 organelle
GO:0043229 0.80 intracellular organelle
GO:0044444 0.73 cytoplasmic part
GO:0043227 0.71 membrane-bounded organelle
GO:0043231 0.67 intracellular membrane-bounded organelle
GO:0005737 0.62 cytoplasm
GO:0044422 0.61 organelle part
GO:0044446 0.60 intracellular organelle part
GO:0032991 0.60 protein-containing complex
GO:0005634 0.55 nucleus
$$$$
A8MYV0-1 Protein serine/threonine kinase acting on microtubule
==== MF ====
* GO:0004674 0.69 protein serine/threonine kinase activity
==== BP ====
GO:0009987 0.99 cellular process
GO:0032502 0.73 developmental process
GO:0016043 0.71 cellular component organization
GO:0019538 0.67 protein metabolic process
GO:0065007 0.61 biological regulation
GO:0048869 0.61 cellular developmental process
GO:0050789 0.60 regulation of biological process
GO:0050794 0.59 regulation of cellular process
GO:0048856 0.59 anatomical structure development
GO:0044237 0.54 cellular metabolic process
GO:0022607 0.54 cellular component assembly
GO:0006996 0.54 organelle organization
* GO:0006468 0.53 protein phosphorylation
GO:0050896 0.48 response to stimulus
* GO:0007017 0.48 microtubule-based process
* GO:0000226 0.47 microtubule cytoskeleton organization
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.96 intracellular part
GO:0043226 0.74 organelle
GO:0044444 0.57 cytoplasmic part
GO:0043229 0.52 intracellular organelle
* GO:0005856 0.47 cytoskeleton
$$$$
O14894-1
==== MF ====
==== BP ====
GO:0050789 0.89 regulation of biological process
GO:0050794 0.87 regulation of cellular process
GO:0009987 0.86 cellular process
GO:0032502 0.79 developmental process
GO:0048856 0.76 anatomical structure development
GO:0048519 0.67 negative regulation of biological process
GO:0048523 0.66 negative regulation of cellular process
GO:0048869 0.62 cellular developmental process
GO:0019222 0.52 regulation of metabolic process
GO:0060255 0.50 regulation of macromolecule metabolic process
GO:0031323 0.50 regulation of cellular metabolic process
==== CC ====
GO:0016021 1.00 integral component of membrane
GO:0044464 0.97 cell part
GO:0044424 0.71 intracellular part
GO:0044459 0.58 plasma membrane part
GO:0005887 0.53 integral component of plasma membrane
$$$$
O60829-1 hydratase (hydrolyase) to degrade phosphate containing molecules, such as RNA
==== MF ====
==== BP ====
* GO:0006139 0.50 nucleobase-containing compound metabolic process
==== CC ====
* GO:0000015 1.00 phosphopyruvate hydratase complex
GO:0005737 0.94 cytoplasm
GO:0043226 0.65 organelle
GO:1990904 0.64 ribonucleoprotein complex
GO:0043229 0.58 intracellular organelle
GO:0009986 0.56 cell surface
GO:0043232 0.53 intracellular non-membrane-bounded organelle
* GO:0043186 0.50 P granule
$$$$
O60941-1 cytoskeleton-associated protein, possibly regulating muscle contraction
==== MF ====
* GO:0008092 0.79 cytoskeletal protein binding
* GO:0005198 0.54 structural molecule activity
* GO:0008307 0.49 structural constituent of muscle
==== BP ====
GO:0009987 0.97 cellular process
GO:0065007 0.81 biological regulation
GO:0050789 0.79 regulation of biological process
GO:0016043 0.78 cellular component organization
GO:0032502 0.74 developmental process
GO:0048856 0.65 anatomical structure development
GO:0050794 0.63 regulation of cellular process
GO:0032501 0.62 multicellular organismal process
GO:0006996 0.57 organelle organization
* GO:0061061 0.55 muscle structure development
GO:0048518 0.54 positive regulation of biological process
* GO:0007010 0.49 cytoskeleton organization
GO:0048513 0.47 animal organ development
GO:0003008 0.47 system process
* GO:0030029 0.46 actin filament-based process
==== CC ====
GO:0044424 1.00 intracellular part
GO:0044425 0.92 membrane part
GO:0044459 0.90 plasma membrane part
GO:0032991 0.90 protein-containing complex
GO:0098797 0.82 plasma membrane protein complex
* GO:0016010 0.75 dystrophin-associated glycoprotein complex
GO:0016020 0.69 membrane
GO:0044422 0.68 organelle part
GO:0044444 0.67 cytoplasmic part
GO:0044446 0.63 intracellular organelle part
GO:0043226 0.63 organelle
GO:0005886 0.63 plasma membrane
GO:0043229 0.62 intracellular organelle
GO:0005737 0.53 cytoplasm
GO:0043227 0.51 membrane-bounded organelle
* GO:0042383 0.49 sarcolemma
$$$$
O75363-1 (for CC: neuron part)
==== MF ====
GO:0042803 0.63 protein homodimerization activity
==== BP ====
GO:0009987 0.90 cellular process
GO:0065007 0.81 biological regulation
GO:0050789 0.74 regulation of biological process
GO:0050794 0.72 regulation of cellular process
GO:0008152 0.70 metabolic process
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.92 intracellular part
GO:0043226 0.74 organelle
GO:0043227 0.70 membrane-bounded organelle
GO:0044444 0.58 cytoplasmic part
GO:0043229 0.52 intracellular organelle
GO:0044421 0.47 extracellular region part
GO:0043231 0.46 intracellular membrane-bounded organelle
GO:0044422 0.43 organelle part
* GO:0097458 0.42 neuron part
$$$$
O75677-1 ubiquitin-protein transferase activity
==== MF ====
GO:0140096 0.80 catalytic activity, acting on a protein
GO:0016740 0.80 transferase activity
* GO:0004842 0.78 ubiquitin-protein transferase activity
==== BP ====
GO:0009987 0.97 cellular process
GO:0065007 0.93 biological regulation
GO:0050789 0.92 regulation of biological process
GO:0050794 0.88 regulation of cellular process
GO:0008152 0.83 metabolic process
GO:0044237 0.82 cellular metabolic process
GO:0071704 0.75 organic substance metabolic process
GO:0044238 0.74 primary metabolic process
GO:0006807 0.72 nitrogen compound metabolic process
GO:0048583 0.71 regulation of response to stimulus
GO:0043170 0.70 macromolecule metabolic process
GO:0048519 0.69 negative regulation of biological process
GO:0048518 0.69 positive regulation of biological process
GO:0019222 0.69 regulation of metabolic process
GO:0050896 0.68 response to stimulus
GO:0016043 0.68 cellular component organization
GO:1901564 0.67 organonitrogen compound metabolic process
GO:0031323 0.67 regulation of cellular metabolic process
GO:0060255 0.66 regulation of macromolecule metabolic process
GO:0044260 0.66 cellular macromolecule metabolic process
GO:0080090 0.65 regulation of primary metabolic process
GO:0051171 0.65 regulation of nitrogen compound metabolic process
GO:0023051 0.65 regulation of signaling
GO:0019538 0.65 protein metabolic process
GO:0010646 0.65 regulation of cell communication
GO:0009966 0.64 regulation of signal transduction
GO:0002376 0.63 immune system process
GO:0048523 0.61 negative regulation of cellular process
GO:0006950 0.61 response to stress
GO:0048522 0.59 positive regulation of cellular process
GO:0044267 0.59 cellular protein metabolic process
GO:0043412 0.59 macromolecule modification
GO:0006464 0.58 cellular protein modification process
GO:0010468 0.56 regulation of gene expression
* GO:0016567 0.55 protein ubiquitination
GO:0051252 0.54 regulation of RNA metabolic process
GO:0031326 0.54 regulation of cellular biosynthetic process
GO:0006355 0.53 regulation of transcription, DNA-templated
GO:0006952 0.51 defense response
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.99 intracellular part
GO:0043226 0.81 organelle
GO:0043229 0.79 intracellular organelle
GO:0044444 0.73 cytoplasmic part
GO:0043227 0.68 membrane-bounded organelle
GO:0032991 0.66 protein-containing complex
GO:0043231 0.64 intracellular membrane-bounded organelle
GO:0044422 0.63 organelle part
GO:0044446 0.62 intracellular organelle part
GO:0005737 0.59 cytoplasm
GO:0005634 0.54 nucleus
$$$$
O95801-1 heat shock protein binding
==== MF ====
GO:0140096 0.59 catalytic activity, acting on a protein
* GO:0031072 0.48 heat shock protein binding
==== BP ====
GO:0009987 0.96 cellular process
GO:0065007 0.68 biological regulation
GO:0050789 0.65 regulation of biological process
GO:0008152 0.64 metabolic process
GO:0071704 0.63 organic substance metabolic process
GO:0044237 0.63 cellular metabolic process
GO:0044238 0.62 primary metabolic process
GO:0050794 0.59 regulation of cellular process
GO:0071840 0.56 cellular component organization or biogenesis
GO:0016043 0.55 cellular component organization
GO:0006807 0.52 nitrogen compound metabolic process
GO:0043170 0.50 macromolecule metabolic process
==== CC ====
GO:0044424 1.00 intracellular part
GO:0044444 0.93 cytoplasmic part
GO:0005737 0.84 cytoplasm
GO:0043226 0.79 organelle
GO:0043229 0.77 intracellular organelle
GO:0016020 0.74 membrane
GO:0043227 0.73 membrane-bounded organelle
GO:0044422 0.72 organelle part
GO:0044446 0.71 intracellular organelle part
GO:0043231 0.69 intracellular membrane-bounded organelle
GO:0005829 0.66 cytosol
GO:0032991 0.64 protein-containing complex
GO:0005634 0.59 nucleus
GO:0005886 0.56 plasma membrane
GO:0044428 0.51 nuclear part
$$$$
P0C870-1 histone demethylation
==== MF ====
GO:0016491 0.79 oxidoreductase activity
GO:0016705 0.78 oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen
GO:0140096 0.77 catalytic activity, acting on a protein
* GO:0016706 0.76 2-oxoglutarate-dependent dioxygenase activity
GO:1901363 0.69 heterocyclic compound binding
GO:0097159 0.69 organic cyclic compound binding
* GO:0032452 0.63 histone demethylase activity
GO:0043167 0.56 ion binding
* GO:0003682 0.51 chromatin binding
==== BP ====
GO:0009987 1.00 cellular process
GO:0008152 0.98 metabolic process
GO:0071704 0.97 organic substance metabolic process
GO:0044238 0.97 primary metabolic process
GO:0044237 0.97 cellular metabolic process
GO:1901564 0.96 organonitrogen compound metabolic process
GO:0043412 0.95 macromolecule modification
GO:0044260 0.84 cellular macromolecule metabolic process
GO:0065007 0.78 biological regulation
GO:0050789 0.77 regulation of biological process
GO:0006464 0.76 cellular protein modification process
GO:0050794 0.71 regulation of cellular process
GO:0019222 0.64 regulation of metabolic process
GO:0060255 0.63 regulation of macromolecule metabolic process
GO:0080090 0.62 regulation of primary metabolic process
GO:0051171 0.62 regulation of nitrogen compound metabolic process
GO:0031323 0.62 regulation of cellular metabolic process
GO:1901576 0.61 organic substance biosynthetic process
GO:0016043 0.60 cellular component organization
GO:0019219 0.59 regulation of nucleobase-containing compound metabolic process
GO:0010468 0.59 regulation of gene expression
GO:0051252 0.58 regulation of RNA metabolic process
GO:1901360 0.56 organic cyclic compound metabolic process
GO:0051179 0.56 localization
GO:0046483 0.56 heterocycle metabolic process
GO:0006725 0.56 cellular aromatic compound metabolic process
GO:1901137 0.55 carbohydrate derivative biosynthetic process
GO:0050896 0.55 response to stimulus
GO:0006810 0.55 transport
GO:0006139 0.55 nucleobase-containing compound metabolic process
GO:0031590 0.54 wybutosine metabolic process
GO:0009451 0.54 RNA modification
GO:0008033 0.54 tRNA processing
* GO:0016570 0.53 histone modification
GO:0051716 0.52 cellular response to stimulus
GO:0042221 0.50 response to chemical
==== CC ====
GO:0044424 1.00 intracellular part
GO:0043229 0.89 intracellular organelle
GO:0043227 0.88 membrane-bounded organelle
GO:0043231 0.87 intracellular membrane-bounded organelle
GO:0005634 0.77 nucleus
GO:0044446 0.72 intracellular organelle part
GO:0044444 0.65 cytoplasmic part
GO:0032991 0.63 protein-containing complex
GO:0042597 0.61 periplasmic space
GO:1902494 0.60 catalytic complex
GO:0044428 0.57 nuclear part
$$$$
P0DPB3-1 DNA binding
==== MF ====
GO:1901363 0.61 heterocyclic compound binding
GO:0097159 0.61 organic cyclic compound binding
* GO:0003677 0.48 DNA binding
==== BP ====
GO:0009987 1.00 cellular process
GO:0050789 0.97 regulation of biological process
GO:0050794 0.93 regulation of cellular process
GO:0048518 0.72 positive regulation of biological process
GO:0048522 0.71 positive regulation of cellular process
GO:0032502 0.64 developmental process
GO:0016043 0.63 cellular component organization
GO:0060255 0.62 regulation of macromolecule metabolic process
GO:0051128 0.61 regulation of cellular component organization
GO:0050793 0.59 regulation of developmental process
GO:0009893 0.59 positive regulation of metabolic process
GO:0010604 0.58 positive regulation of macromolecule metabolic process
GO:0080090 0.57 regulation of primary metabolic process
GO:0031323 0.57 regulation of cellular metabolic process
GO:0051171 0.56 regulation of nitrogen compound metabolic process
GO:0051173 0.53 positive regulation of nitrogen compound metabolic process
GO:0031325 0.53 positive regulation of cellular metabolic process
GO:0008152 0.53 metabolic process
GO:0071704 0.52 organic substance metabolic process
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.96 intracellular part
GO:0005737 0.74 cytoplasm
GO:0044444 0.59 cytoplasmic part
GO:0044422 0.58 organelle part
GO:0016020 0.57 membrane
GO:0043226 0.54 organelle
GO:0043229 0.51 intracellular organelle
GO:0030054 0.50 cell junction
$$$$
P56378-1 ATPase or ATP synthase in mitochondrion
==== MF ====
GO:0016787 0.55 hydrolase activity
* GO:0016887 0.49 ATPase activity
==== BP ====
GO:0009987 0.83 cellular process
GO:0008152 0.72 metabolic process
GO:0044237 0.71 cellular metabolic process
GO:0071704 0.68 organic substance metabolic process
GO:0044238 0.68 primary metabolic process
GO:0006807 0.67 nitrogen compound metabolic process
GO:0034641 0.60 cellular nitrogen compound metabolic process
GO:1901360 0.59 organic cyclic compound metabolic process
GO:0006139 0.58 nucleobase-containing compound metabolic process
GO:1901564 0.55 organonitrogen compound metabolic process
GO:0006810 0.54 transport
==== CC ====
GO:0005753 1.00 mitochondrial proton-transporting ATP synthase complex
GO:0043229 0.80 intracellular organelle
GO:0043227 0.78 membrane-bounded organelle
GO:0043231 0.77 intracellular membrane-bounded organelle
* GO:0005739 0.70 mitochondrion
$$$$
P60827-1 signaling receptor binding
==== MF ====
GO:0005102 0.41 signaling receptor binding
==== BP ====
GO:0009987 0.93 cellular process
GO:0065007 0.85 biological regulation
GO:0050789 0.82 regulation of biological process
GO:0048518 0.67 positive regulation of biological process
GO:0032502 0.65 developmental process
GO:0050896 0.62 response to stimulus
GO:0050794 0.62 regulation of cellular process
GO:0048583 0.62 regulation of response to stimulus
GO:0016043 0.61 cellular component organization
GO:0008152 0.61 metabolic process
GO:0071704 0.56 organic substance metabolic process
GO:0044238 0.54 primary metabolic process
GO:0048584 0.53 positive regulation of response to stimulus
==== CC ====
GO:0016020 1.00 membrane
GO:0044464 0.83 cell part
GO:0044421 0.77 extracellular region part
GO:0044424 0.65 intracellular part
GO:0005615 0.58 extracellular space
GO:0043226 0.53 organelle
GO:0043227 0.52 membrane-bounded organelle
GO:0005576 0.50 extracellular region
$$$$
P84157-1
==== MF ====
==== BP ====
GO:0009987 0.88 cellular process
GO:0065007 0.71 biological regulation
GO:0050789 0.66 regulation of biological process
GO:0008152 0.64 metabolic process
GO:0050794 0.61 regulation of cellular process
GO:0044237 0.61 cellular metabolic process
GO:0071704 0.50 organic substance metabolic process
==== CC ====
GO:0044424 1.00 intracellular part
GO:0044446 0.84 intracellular organelle part
GO:0032991 0.82 protein-containing complex
GO:0043226 0.77 organelle
GO:0043229 0.75 intracellular organelle
GO:0043227 0.73 membrane-bounded organelle
GO:0043231 0.70 intracellular membrane-bounded organelle
GO:0005634 0.61 nucleus
GO:0044444 0.56 cytoplasmic part
GO:0005737 0.53 cytoplasm
GO:0044428 0.51 nuclear part
$$$$
Q494U1-1 transmembrane transport of small molecules, such as nucleotide
==== MF ====
GO:0022857 0.93 transmembrane transporter activity
GO:0015075 0.76 ion transmembrane transporter activity
GO:0008509 0.75 anion transmembrane transporter activity
GO:0008514 0.72 organic anion transmembrane transporter activity
GO:0015605 0.63 organophosphate ester transmembrane transporter activity
GO:0015318 0.63 inorganic molecular entity transmembrane transporter activity
* GO:0008028 0.59 monocarboxylic acid transmembrane transporter activity
==== BP ====
GO:0006810 0.96 transport
GO:0071702 0.93 organic substance transport
GO:0055085 0.87 transmembrane transport
GO:0009987 0.86 cellular process
GO:0006811 0.78 ion transport
GO:0034220 0.77 ion transmembrane transport
GO:0015711 0.77 organic anion transport
GO:0098656 0.76 anion transmembrane transport
GO:0071705 0.71 nitrogen compound transport
GO:1901264 0.68 carbohydrate derivative transport
GO:0065007 0.62 biological regulation
GO:0051641 0.57 cellular localization
GO:0050789 0.57 regulation of biological process
GO:0050794 0.56 regulation of cellular process
GO:0046907 0.56 intracellular transport
GO:0015931 0.55 nucleobase-containing compound transport
GO:0007017 0.55 microtubule-based process
GO:0006928 0.55 movement of cell or subcellular component
GO:0046903 0.53 secretion
GO:0030705 0.53 cytoskeleton-dependent intracellular transport
* GO:0015780 0.53 nucleotide-sugar transmembrane transport
==== CC ====
GO:0044424 1.00 intracellular part
GO:0044444 0.99 cytoplasmic part
GO:0043226 0.97 organelle
GO:0043231 0.96 intracellular membrane-bounded organelle
GO:0044446 0.84 intracellular organelle part
GO:0005737 0.71 cytoplasm
GO:0032991 0.69 protein-containing complex
* GO:0016020 0.62 membrane
GO:0005794 0.58 Golgi apparatus
GO:0005634 0.53 nucleus
GO:0044428 0.52 nuclear part
$$$$
Q5T0D9-1 phosphatidylinositol-4-phosphate phosphatase
==== MF ====
GO:0016787 0.93 hydrolase activity
GO:0042803 0.92 protein homodimerization activity
GO:0016791 0.92 phosphatase activity
* GO:0034596 0.70 phosphatidylinositol phosphate 4-phosphatase activity
* GO:0052833 0.68 inositol monophosphate 4-phosphatase activity
==== BP ====
GO:0065007 0.96 biological regulation
GO:0050794 0.95 regulation of cellular process
GO:0006810 0.94 transport
GO:0051049 0.93 regulation of transport
GO:2001135 0.92 regulation of endocytic recycling
GO:0072583 0.92 clathrin-dependent endocytosis
GO:0009987 0.91 cellular process
GO:0008152 0.81 metabolic process
GO:0071704 0.80 organic substance metabolic process
GO:0044238 0.80 primary metabolic process
GO:0044237 0.80 cellular metabolic process
GO:0019222 0.76 regulation of metabolic process
GO:0060255 0.75 regulation of macromolecule metabolic process
GO:0050896 0.75 response to stimulus
GO:0048523 0.75 negative regulation of cellular process
GO:0048518 0.75 positive regulation of biological process
GO:0080090 0.74 regulation of primary metabolic process
GO:0051171 0.74 regulation of nitrogen compound metabolic process
GO:0048522 0.74 positive regulation of cellular process
GO:0044248 0.74 cellular catabolic process
GO:0031323 0.74 regulation of cellular metabolic process
GO:1901575 0.73 organic substance catabolic process
GO:0048583 0.73 regulation of response to stimulus
GO:0032501 0.73 multicellular organismal process
GO:0031325 0.73 positive regulation of cellular metabolic process
GO:0023051 0.73 regulation of signaling
GO:0010646 0.73 regulation of cell communication
GO:0010604 0.73 positive regulation of macromolecule metabolic process
GO:0006950 0.73 response to stress
GO:0051246 0.72 regulation of protein metabolic process
GO:0051239 0.72 regulation of multicellular organismal process
GO:0051172 0.72 negative regulation of nitrogen compound metabolic process
GO:0050793 0.72 regulation of developmental process
GO:0031324 0.72 negative regulation of cellular metabolic process
GO:0023056 0.72 positive regulation of signaling
GO:0019637 0.72 organophosphate metabolic process
GO:0010605 0.72 negative regulation of macromolecule metabolic process
GO:0009966 0.72 regulation of signal transduction
GO:0006796 0.72 phosphate-containing compound metabolic process
GO:2000145 0.71 regulation of cell motility
GO:1902531 0.71 regulation of intracellular signal transduction
GO:0080134 0.71 regulation of response to stress
GO:0051241 0.71 negative regulation of multicellular organismal process
GO:0051129 0.71 negative regulation of cellular component organization
GO:0050767 0.71 regulation of neurogenesis
GO:0045596 0.71 negative regulation of cell differentiation
GO:0044255 0.71 cellular lipid metabolic process
GO:0032101 0.71 regulation of response to external stimulus
GO:0009968 0.71 negative regulation of signal transduction
GO:0003008 0.71 system process
GO:0001932 0.71 regulation of protein phosphorylation
GO:0048681 0.70 negative regulation of axon regeneration
* GO:0048015 0.70 phosphatidylinositol-mediated signaling
* GO:0046856 0.70 phosphatidylinositol dephosphorylation
GO:0042532 0.70 negative regulation of tyrosine phosphorylation of STAT protein
* GO:0031161 0.70 phosphatidylinositol catabolic process
GO:0014898 0.70 cardiac muscle hypertrophy in response to stress
GO:0001921 0.70 positive regulation of receptor recycling
==== CC ====
GO:0044424 1.00 intracellular part
GO:0005737 0.94 cytoplasm
GO:0044444 0.63 cytoplasmic part
GO:0043226 0.57 organelle
GO:0043229 0.54 intracellular organelle
GO:0043227 0.54 membrane-bounded organelle
$$$$
Q5VTQ0-1 protein ubiquitination regulation
==== MF ====
* GO:0019899 0.52 enzyme binding
==== BP ====
GO:0009987 0.96 cellular process
GO:0065007 0.81 biological regulation
GO:0050789 0.79 regulation of biological process
GO:0050794 0.76 regulation of cellular process
GO:0008152 0.73 metabolic process
GO:0071704 0.72 organic substance metabolic process
GO:0044237 0.72 cellular metabolic process
GO:0044238 0.71 primary metabolic process
GO:0006807 0.69 nitrogen compound metabolic process
GO:0043170 0.67 macromolecule metabolic process
GO:1901564 0.65 organonitrogen compound metabolic process
GO:0044260 0.64 cellular macromolecule metabolic process
GO:0019538 0.62 protein metabolic process
GO:0006464 0.60 cellular protein modification process
GO:1901575 0.53 organic substance catabolic process
GO:0044248 0.53 cellular catabolic process
GO:0051128 0.52 regulation of cellular component organization
* GO:0006508 0.52 proteolysis
GO:0044265 0.51 cellular macromolecule catabolic process
GO:0033043 0.50 regulation of organelle organization
* GO:0016567 0.50 protein ubiquitination
* GO:0051603 0.49 proteolysis involved in cellular protein catabolic process
* GO:0043632 0.49 modification-dependent macromolecule catabolic process
==== CC ====
GO:0044424 1.00 intracellular part
GO:0044444 0.91 cytoplasmic part
GO:0032991 0.91 protein-containing complex
GO:0043226 0.88 organelle
GO:0043229 0.84 intracellular organelle
GO:0043227 0.83 membrane-bounded organelle
GO:0005737 0.82 cytoplasm
GO:0043231 0.81 intracellular membrane-bounded organelle
GO:0044422 0.78 organelle part
GO:0044446 0.77 intracellular organelle part
GO:0005634 0.74 nucleus
GO:1902494 0.71 catalytic complex
GO:0044428 0.68 nuclear part
GO:0005829 0.64 cytosol
GO:1990234 0.53 transferase complex
$$$$
Q6AI39-1 sodium:potassium ion transporter
==== MF ====
GO:0097159 0.79 organic cyclic compound binding
GO:1901363 0.78 heterocyclic compound binding
GO:0043167 0.65 ion binding
GO:0016787 0.63 hydrolase activity
GO:0043168 0.61 anion binding
GO:0097367 0.59 carbohydrate derivative binding
GO:0036094 0.59 small molecule binding
GO:0016462 0.59 pyrophosphatase activity
GO:0017111 0.58 nucleoside-triphosphatase activity
GO:0016887 0.57 ATPase activity
GO:0042623 0.56 ATPase activity, coupled
GO:0022857 0.56 transmembrane transporter activity
GO:0046873 0.55 metal ion transmembrane transporter activity
* GO:0005391 0.54 sodium:potassium-exchanging ATPase activity
==== BP ====
GO:0009987 0.95 cellular process
GO:0065007 0.90 biological regulation
GO:0050789 0.86 regulation of biological process
GO:0050794 0.75 regulation of cellular process
GO:0051179 0.67 localization
GO:0006810 0.63 transport
GO:0019222 0.58 regulation of metabolic process
GO:0065008 0.57 regulation of biological quality
GO:0060255 0.55 regulation of macromolecule metabolic process
GO:0031323 0.55 regulation of cellular metabolic process
GO:0006811 0.55 ion transport
* GO:0010248 0.54 establishment or maintenance of transmembrane electrochemical gradient
GO:0080090 0.53 regulation of primary metabolic process
GO:0051171 0.52 regulation of nitrogen compound metabolic process
GO:0010468 0.52 regulation of gene expression
GO:0071704 0.50 organic substance metabolic process
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.96 intracellular part
GO:0044425 0.85 membrane part
GO:0016021 0.81 integral component of membrane
GO:0043226 0.75 organelle
GO:0016020 0.75 membrane
GO:0044422 0.74 organelle part
GO:0043229 0.73 intracellular organelle
GO:0044446 0.72 intracellular organelle part
GO:0043227 0.69 membrane-bounded organelle
GO:0043231 0.65 intracellular membrane-bounded organelle
GO:0032991 0.65 protein-containing complex
GO:0044444 0.59 cytoplasmic part
* GO:0005886 0.55 plasma membrane
GO:0005634 0.55 nucleus
GO:0044428 0.52 nuclear part
$$$$
Q6ZNE9-2 regulation of protein folding
==== MF ====
* GO:0051082 0.44 unfolded protein binding
* GO:0044183 0.44 protein binding involved in protein folding
GO:0019899 0.42 enzyme binding
GO:0051020 0.40 GTPase binding
==== BP ====
GO:0009987 0.96 cellular process
GO:0065007 0.78 biological regulation
GO:0071704 0.76 organic substance metabolic process
GO:0044238 0.76 primary metabolic process
GO:0044237 0.76 cellular metabolic process
GO:0050794 0.75 regulation of cellular process
GO:0006807 0.75 nitrogen compound metabolic process
GO:0006139 0.74 nucleobase-containing compound metabolic process
GO:0034654 0.71 nucleobase-containing compound biosynthetic process
GO:0051179 0.69 localization
GO:0051128 0.68 regulation of cellular component organization
GO:0006810 0.68 transport
GO:0055086 0.63 nucleobase-containing small molecule metabolic process
GO:0051641 0.63 cellular localization
GO:0006793 0.63 phosphorus metabolic process
GO:0046907 0.57 intracellular transport
GO:0031323 0.54 regulation of cellular metabolic process
GO:1901135 0.53 carbohydrate derivative metabolic process
* GO:0061077 0.51 chaperone-mediated protein folding
* GO:0006458 0.51 'de novo' protein folding
GO:0033043 0.49 regulation of organelle organization
GO:0051640 0.46 organelle localization
GO:0048518 0.46 positive regulation of biological process
GO:0051656 0.45 establishment of organelle localization
GO:0048522 0.45 positive regulation of cellular process
GO:1901564 0.43 organonitrogen compound metabolic process
GO:0051130 0.43 positive regulation of cellular component organization
GO:1901137 0.41 carbohydrate derivative biosynthetic process
==== CC ====
GO:0044446 1.00 intracellular organelle part
GO:0043231 1.00 intracellular membrane-bounded organelle
GO:0044444 0.98 cytoplasmic part
GO:0005737 0.76 cytoplasm
GO:0005829 0.72 cytosol
GO:0032991 0.68 protein-containing complex
GO:0005634 0.66 nucleus
GO:0016020 0.58 membrane
GO:0031982 0.54 vesicle
GO:0031410 0.48 cytoplasmic vesicle
GO:0005768 0.46 endosome
GO:0044428 0.45 nuclear part
$$$$
Q6ZW76-1 transferase acting on a protein
==== MF ====
* GO:0140096 0.52 catalytic activity, acting on a protein
* GO:0016740 0.51 transferase activity
==== BP ====
GO:0009987 0.95 cellular process
GO:0065007 0.86 biological regulation
GO:0050789 0.83 regulation of biological process
GO:0050794 0.81 regulation of cellular process
GO:0044237 0.58 cellular metabolic process
GO:0071704 0.57 organic substance metabolic process
GO:0044238 0.56 primary metabolic process
GO:0006807 0.54 nitrogen compound metabolic process
GO:0016043 0.52 cellular component organization
GO:0019222 0.51 regulation of metabolic process
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.98 intracellular part
GO:0043226 0.86 organelle
GO:0043229 0.85 intracellular organelle
GO:0043227 0.82 membrane-bounded organelle
GO:0043231 0.75 intracellular membrane-bounded organelle
GO:0044444 0.74 cytoplasmic part
GO:0005737 0.70 cytoplasm
GO:0016020 0.69 membrane
GO:0005634 0.64 nucleus
GO:0044425 0.63 membrane part
GO:0044422 0.62 organelle part
GO:0044446 0.57 intracellular organelle part
GO:0016021 0.57 integral component of membrane
GO:0032991 0.56 protein-containing complex
$$$$
Q70Z53-1 cytoskelton organization and cytoskelton based transport
==== MF ====
GO:0016787 0.58 hydrolase activity
* GO:0045503 0.48 dynein light chain binding
==== BP ====
GO:0009987 0.95 cellular process
GO:0016043 0.78 cellular component organization
GO:0032502 0.77 developmental process
GO:0006996 0.76 organelle organization
GO:0009653 0.75 anatomical structure morphogenesis
GO:0048598 0.74 embryonic morphogenesis
GO:0008152 0.66 metabolic process
GO:0071704 0.60 organic substance metabolic process
GO:0065007 0.60 biological regulation
GO:0044238 0.60 primary metabolic process
GO:0044237 0.59 cellular metabolic process
GO:0006807 0.59 nitrogen compound metabolic process
GO:0050789 0.58 regulation of biological process
GO:0050794 0.57 regulation of cellular process
GO:0043170 0.56 macromolecule metabolic process
GO:0022607 0.53 cellular component assembly
GO:0034641 0.52 cellular nitrogen compound metabolic process
GO:0120036 0.51 plasma membrane bounded cell projection organization
GO:0070925 0.51 organelle assembly
GO:0006139 0.51 nucleobase-containing compound metabolic process
GO:0090304 0.50 nucleic acid metabolic process
* GO:0060271 0.50 cilium assembly
* GO:0048704 0.50 embryonic skeletal system morphogenesis
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.99 intracellular part
GO:0044425 0.80 membrane part
GO:0043226 0.79 organelle
GO:0016021 0.78 integral component of membrane
GO:0043227 0.77 membrane-bounded organelle
GO:0016020 0.76 membrane
GO:0044422 0.75 organelle part
GO:0044446 0.74 intracellular organelle part
GO:0032991 0.70 protein-containing complex
GO:0044444 0.67 cytoplasmic part
GO:0043229 0.58 intracellular organelle
GO:1902494 0.56 catalytic complex
GO:0043231 0.54 intracellular membrane-bounded organelle
GO:0031982 0.52 vesicle
GO:0120038 0.51 plasma membrane bounded cell projection part
* GO:0044430 0.50 cytoskeletal part
GO:0044421 0.50 extracellular region part
GO:0120025 0.49 plasma membrane bounded cell projection
GO:0005615 0.49 extracellular space
* GO:0000922 0.49 spindle pole
GO:0097546 0.48 ciliary base
GO:0070062 0.48 extracellular exosome
* GO:0031021 0.48 interphase microtubule organizing center
GO:0005929 0.48 cilium
* GO:0005868 0.48 cytoplasmic dynein complex
* GO:0005813 0.48 centrosome
* GO:0000242 0.48 pericentriolar material
$$$$
Q7Z5A7-1 regulation of microglial cell activation
==== MF ====
==== BP ====
GO:0050789 1.00 regulation of biological process
GO:0050794 0.85 regulation of cellular process
GO:0009987 0.84 cellular process
GO:0048522 0.75 positive regulation of cellular process
GO:0048583 0.73 regulation of response to stimulus
GO:0048523 0.72 negative regulation of cellular process
GO:0048584 0.71 positive regulation of response to stimulus
GO:0048585 0.67 negative regulation of response to stimulus
* GO:0031347 0.67 regulation of defense response
* GO:0002682 0.67 regulation of immune system process
* GO:1903980 0.66 positive regulation of microglial cell activation
* GO:1903979 0.66 negative regulation of microglial cell activation
==== CC ====
GO:0044464 0.95 cell part
GO:0044424 0.85 intracellular part
GO:0044421 0.74 extracellular region part
GO:0005615 0.73 extracellular space
GO:0043226 0.67 organelle
GO:0043229 0.66 intracellular organelle
GO:0044444 0.62 cytoplasmic part
GO:0043227 0.62 membrane-bounded organelle
GO:0043231 0.60 intracellular membrane-bounded organelle
GO:0005783 0.50 endoplasmic reticulum
$$$$
Q8IUW5-1 regulation of apoptosis through TNF
==== MF ====
* GO:0005031 0.40 tumor necrosis factor-activated receptor activity
==== BP ====
GO:0050789 1.00 regulation of biological process
GO:0009987 0.70 cellular process
GO:0050794 0.65 regulation of cellular process
GO:0048518 0.64 positive regulation of biological process
* GO:0097190 0.51 apoptotic signaling pathway
* GO:0042981 0.51 regulation of apoptotic process
* GO:0042127 0.51 regulation of cell proliferation
GO:0032496 0.51 response to lipopolysaccharide
GO:0007275 0.51 multicellular organism development
* GO:0006955 0.51 immune response
* GO:0006954 0.51 inflammatory response
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.80 intracellular part
GO:0043229 0.76 intracellular organelle
$$$$
Q8IUY3-1 binding of GTPase from Ras superfamily
==== MF ====
GO:0098772 0.64 molecular function regulator
* GO:0017016 0.63 Ras GTPase binding
* GO:0005096 0.62 GTPase activator activity
* GO:0017137 0.61 Rab GTPase binding
==== BP ====
GO:0065007 0.85 biological regulation
GO:0051179 0.84 localization
GO:0051641 0.83 cellular localization
GO:0050794 0.82 regulation of cellular process
GO:0071702 0.81 organic substance transport
GO:0046907 0.81 intracellular transport
GO:0050790 0.79 regulation of catalytic activity
GO:0043085 0.78 positive regulation of catalytic activity
GO:0051336 0.77 regulation of hydrolase activity
GO:0051345 0.76 positive regulation of hydrolase activity
GO:0032879 0.76 regulation of localization
GO:0051128 0.74 regulation of cellular component organization
* GO:0043087 0.74 regulation of GTPase activity
* GO:0090630 0.73 activation of GTPase activity
GO:0031338 0.73 regulation of vesicle fusion
GO:0006886 0.73 intracellular protein transport
==== CC ====
GO:0044464 0.96 cell part
GO:0012505 0.56 endomembrane system
GO:0005622 0.56 intracellular
$$$$
Q8N957-1 nucleotide excision repair
==== MF ====
==== BP ====
GO:0009987 0.97 cellular process
GO:0065007 0.85 biological regulation
GO:0050789 0.83 regulation of biological process
GO:0050794 0.81 regulation of cellular process
GO:0032502 0.77 developmental process
GO:0016043 0.73 cellular component organization
GO:0008152 0.61 metabolic process
GO:0044237 0.59 cellular metabolic process
GO:0071704 0.58 organic substance metabolic process
GO:0044238 0.56 primary metabolic process
GO:0006807 0.56 nitrogen compound metabolic process
GO:0048518 0.55 positive regulation of biological process
GO:0050896 0.54 response to stimulus
GO:0043170 0.53 macromolecule metabolic process
GO:0044260 0.50 cellular macromolecule metabolic process
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.98 intracellular part
GO:0032991 0.86 protein-containing complex
GO:0043226 0.76 organelle
GO:0044422 0.75 organelle part
GO:0043229 0.73 intracellular organelle
GO:0044446 0.70 intracellular organelle part
GO:0044444 0.68 cytoplasmic part
GO:0043227 0.64 membrane-bounded organelle
GO:0043231 0.59 intracellular membrane-bounded organelle
GO:0005737 0.56 cytoplasm
GO:0005634 0.50 nucleus
* GO:1902494 0.48 catalytic complex
* GO:0009380 0.44 excinuclease repair complex
$$$$
Q8NDM7-1
==== MF ====
==== BP ====
GO:0009987 0.98 cellular process
GO:0065007 0.70 biological regulation
GO:0050789 0.68 regulation of biological process
GO:0050794 0.66 regulation of cellular process
GO:0071840 0.61 cellular component organization or biogenesis
GO:0016043 0.60 cellular component organization
GO:0044237 0.50 cellular metabolic process
==== CC ====
GO:0044464 0.99 cell part
GO:0044424 0.98 intracellular part
GO:0044446 0.81 intracellular organelle part
GO:0043226 0.74 organelle
GO:0043229 0.68 intracellular organelle
GO:0044444 0.65 cytoplasmic part
GO:0043227 0.63 membrane-bounded organelle
GO:0032991 0.63 protein-containing complex
GO:0043231 0.54 intracellular membrane-bounded organelle
$$$$
Q8TDG2-1 regulation of chromosome organization either though histone acetylation or binding of cytoskelton used in chromsome segregation
==== MF ====
GO:0005198 0.90 structural molecule activity
GO:1901363 0.89 heterocyclic compound binding
GO:0097159 0.89 organic cyclic compound binding
GO:0005200 0.89 structural constituent of cytoskeleton
GO:0043167 0.85 ion binding
GO:0035639 0.84 purine ribonucleoside triphosphate binding
GO:0032555 0.84 purine ribonucleotide binding
GO:0016740 0.66 transferase activity
* GO:0004402 0.61 histone acetyltransferase activity
* GO:0008092 0.59 cytoskeletal protein binding
==== BP ====
GO:0065007 0.96 biological regulation
GO:0050789 0.95 regulation of biological process
GO:0009987 0.90 cellular process
GO:0060255 0.89 regulation of macromolecule metabolic process
GO:0048518 0.89 positive regulation of biological process
GO:0010468 0.88 regulation of gene expression
GO:0009893 0.85 positive regulation of metabolic process
GO:0016043 0.78 cellular component organization
GO:0010604 0.78 positive regulation of macromolecule metabolic process
GO:0010628 0.77 positive regulation of gene expression
GO:0032501 0.76 multicellular organismal process
GO:0050794 0.74 regulation of cellular process
GO:0090131 0.71 mesenchyme migration
GO:0032502 0.71 developmental process
GO:0044237 0.70 cellular metabolic process
GO:0071704 0.69 organic substance metabolic process
GO:0044238 0.69 primary metabolic process
GO:0006996 0.69 organelle organization
GO:0050896 0.68 response to stimulus
GO:0006807 0.68 nitrogen compound metabolic process
GO:1901564 0.67 organonitrogen compound metabolic process
GO:0044260 0.67 cellular macromolecule metabolic process
GO:0022607 0.66 cellular component assembly
GO:0051716 0.65 cellular response to stimulus
GO:0048869 0.65 cellular developmental process
GO:0044267 0.65 cellular protein metabolic process
GO:0043412 0.65 macromolecule modification
GO:0042221 0.64 response to chemical
GO:0006950 0.64 response to stress
GO:0006464 0.64 cellular protein modification process
* GO:0006325 0.64 chromatin organization
GO:1901360 0.63 organic cyclic compound metabolic process
GO:0034641 0.63 cellular nitrogen compound metabolic process
GO:0022402 0.63 cell cycle process
GO:0070887 0.62 cellular response to chemical stimulus
GO:0033554 0.62 cellular response to stress
GO:0022414 0.62 reproductive process
GO:0006139 0.62 nucleobase-containing compound metabolic process
GO:0090304 0.61 nucleic acid metabolic process
GO:0048646 0.61 anatomical structure formation involved in morphogenesis
* GO:0018193 0.61 peptidyl-amino acid modification
* GO:0016570 0.61 histone modification
GO:0003006 0.61 developmental process involved in reproduction
GO:1903046 0.60 meiotic cell cycle process
GO:0032506 0.60 cytokinetic process
* GO:0016573 0.60 histone acetylation
GO:0010927 0.60 cellular component assembly involved in morphogenesis
* GO:0006281 0.60 DNA repair
GO:0034599 0.59 cellular response to oxidative stress
GO:0030476 0.59 ascospore wall assembly
GO:0001300 0.59 chronological cell aging
* GO:0000916 0.59 actomyosin contractile ring contraction
GO:0000011 0.59 vacuole inheritance
GO:0080090 0.57 regulation of primary metabolic process
GO:0051171 0.57 regulation of nitrogen compound metabolic process
GO:0031323 0.57 regulation of cellular metabolic process
GO:0031326 0.54 regulation of cellular biosynthetic process
GO:0019219 0.54 regulation of nucleobase-containing compound metabolic process
GO:0010556 0.54 regulation of macromolecule biosynthetic process
GO:0048522 0.53 positive regulation of cellular process
GO:0048519 0.53 negative regulation of biological process
* GO:0006355 0.53 regulation of transcription, DNA-templated
GO:0051179 0.51 localization
GO:0048523 0.51 negative regulation of cellular process
==== CC ====
GO:0044424 1.00 intracellular part
GO:0043226 0.94 organelle
GO:0043229 0.93 intracellular organelle
GO:0044446 0.88 intracellular organelle part
GO:0032991 0.79 protein-containing complex
GO:0044444 0.77 cytoplasmic part
GO:0043227 0.74 membrane-bounded organelle
GO:0005737 0.73 cytoplasm
GO:0043231 0.71 intracellular membrane-bounded organelle
GO:0043232 0.70 intracellular non-membrane-bounded organelle
* GO:0005856 0.66 cytoskeleton
GO:0005634 0.64 nucleus
* GO:0044430 0.60 cytoskeletal part
GO:0042995 0.54 cell projection
GO:0016020 0.53 membrane
$$$$
Q8WTR8-1 anatomical structure morphogenesis
==== MF ====
==== BP ====
GO:0009987 0.98 cellular process
GO:0065007 0.88 biological regulation
GO:0050789 0.86 regulation of biological process
GO:0032502 0.86 developmental process
GO:0050794 0.84 regulation of cellular process
GO:0016043 0.71 cellular component organization
GO:0006928 0.68 movement of cell or subcellular component
* GO:0048856 0.66 anatomical structure development
* GO:0009653 0.63 anatomical structure morphogenesis
GO:0048869 0.62 cellular developmental process
GO:0048518 0.60 positive regulation of biological process
GO:0048522 0.55 positive regulation of cellular process
GO:0051239 0.52 regulation of multicellular organismal process
GO:0050793 0.52 regulation of developmental process
GO:0051128 0.51 regulation of cellular component organization
GO:0048519 0.51 negative regulation of biological process
GO:2000026 0.50 regulation of multicellular organismal development
GO:0048583 0.48 regulation of response to stimulus
* GO:0007411 0.48 axon guidance
GO:0045595 0.47 regulation of cell differentiation
GO:0048523 0.46 negative regulation of cellular process
GO:0022603 0.46 regulation of anatomical structure morphogenesis
GO:0120035 0.45 regulation of plasma membrane bounded cell projection organization
* GO:0051960 0.45 regulation of nervous system development
* GO:0050767 0.44 regulation of neurogenesis
* GO:0045664 0.43 regulation of neuron differentiation
GO:0030030 0.43 cell projection organization
* GO:0010975 0.42 regulation of neuron projection development
* GO:0010769 0.41 regulation of cell morphogenesis involved in differentiation
==== CC ====
GO:0019031 1.00 viral envelope
GO:0044464 0.94 cell part
GO:0044424 0.70 intracellular part
GO:0044421 0.69 extracellular region part
GO:0044425 0.58 membrane part
GO:0062023 0.53 collagen-containing extracellular matrix
GO:0044459 0.52 plasma membrane part
$$$$
Q8WXF5-1 structure component of eye lens
==== MF ====
* GO:0005212 0.95 structural constituent of eye lens
==== BP ====
GO:0032502 0.91 developmental process
GO:0032501 0.86 multicellular organismal process
GO:0003008 0.85 system process
GO:0007601 0.84 visual perception
GO:0009987 0.81 cellular process
GO:0048856 0.77 anatomical structure development
GO:0009653 0.63 anatomical structure morphogenesis
* GO:0048513 0.59 animal organ development
GO:0002009 0.59 morphogenesis of an epithelium
* GO:0001654 0.58 eye development
GO:0065007 0.49 biological regulation
* GO:0002088 0.49 lens development in camera-type eye
==== CC ====
GO:0005737 1.00 cytoplasm
GO:0005634 1.00 nucleus
$$$$
Q96A25-1 lysosomal transport and lysosome organization
==== MF ====
GO:0016787 0.44 hydrolase activity
==== BP ====
GO:0048813 1.00 dendrite morphogenesis
* GO:0032418 1.00 lysosome localization
GO:0006996 0.93 organelle organization
GO:0006810 0.93 transport
* GO:0007041 0.92 lysosomal transport
* GO:0007040 0.92 lysosome organization
GO:0065007 0.64 biological regulation
GO:0050789 0.59 regulation of biological process
GO:0050794 0.57 regulation of cellular process
==== CC ====
GO:0044424 1.00 intracellular part
GO:0044444 0.98 cytoplasmic part
GO:0016020 0.94 membrane
GO:0044446 0.93 intracellular organelle part
GO:0098588 0.87 bounding membrane of organelle
GO:0044437 0.87 vacuolar part
GO:0043226 0.87 organelle
GO:0043229 0.86 intracellular organelle
GO:0005774 0.86 vacuolar membrane
GO:0043227 0.85 membrane-bounded organelle
* GO:0005765 0.85 lysosomal membrane
GO:0043231 0.83 intracellular membrane-bounded organelle
GO:0005773 0.55 vacuole
GO:0031982 0.50 vesicle
$$$$
Q96D15-1 catalytic activity, acting on a protein
==== MF ====
* GO:0140096 0.41 catalytic activity, acting on a protein
==== BP ====
GO:0009987 0.95 cellular process
GO:0065007 0.83 biological regulation
GO:0050789 0.78 regulation of biological process
GO:0050794 0.75 regulation of cellular process
GO:0071704 0.66 organic substance metabolic process
GO:0044237 0.65 cellular metabolic process
GO:0044238 0.64 primary metabolic process
GO:0006807 0.63 nitrogen compound metabolic process
GO:0043170 0.61 macromolecule metabolic process
GO:0044260 0.56 cellular macromolecule metabolic process
GO:1901564 0.54 organonitrogen compound metabolic process
GO:0019538 0.52 protein metabolic process
GO:0043412 0.51 macromolecule modification
==== CC ====
GO:0044424 1.00 intracellular part
GO:0043226 0.94 organelle
GO:0043229 0.93 intracellular organelle
GO:0043227 0.92 membrane-bounded organelle
GO:0044444 0.90 cytoplasmic part
GO:0043231 0.90 intracellular membrane-bounded organelle
GO:0044446 0.82 intracellular organelle part
GO:0016020 0.80 membrane
GO:0005886 0.72 plasma membrane
GO:0005737 0.69 cytoplasm
GO:0044425 0.61 membrane part
GO:0005634 0.61 nucleus
GO:0044459 0.54 plasma membrane part
GO:0005783 0.53 endoplasmic reticulum
$$$$
Q96F63-1 splicesome protein
==== MF ====
==== BP ====
GO:0009987 0.83 cellular process
GO:0008152 0.54 metabolic process
GO:0071704 0.53 organic substance metabolic process
GO:0044238 0.53 primary metabolic process
GO:0044237 0.53 cellular metabolic process
GO:0006807 0.50 nitrogen compound metabolic process
==== CC ====
* GO:0005686 1.00 U2 snRNP
$$$$
Q96II8-1 transferase acting on a protein
==== MF ====
* GO:0140096 0.45 catalytic activity, acting on a protein
* GO:0016740 0.45 transferase activity
==== BP ====
GO:0009987 0.97 cellular process
GO:0065007 0.93 biological regulation
GO:0050789 0.92 regulation of biological process
GO:0050794 0.90 regulation of cellular process
==== CC ====
GO:0044464 1.00 cell part
GO:0016021 1.00 integral component of membrane
GO:0044424 0.94 intracellular part
GO:0016020 0.79 membrane
GO:0005886 0.63 plasma membrane
GO:0005737 0.62 cytoplasm
GO:0043226 0.55 organelle
GO:0044444 0.51 cytoplasmic part
GO:0043229 0.51 intracellular organelle
$$$$
Q96J88-1 cytoskeleton binding
==== MF ====
* GO:0008092 0.43 cytoskeletal protein binding
==== BP ====
GO:0009987 0.95 cellular process
GO:0065007 0.83 biological regulation
GO:0050789 0.80 regulation of biological process
GO:0050794 0.77 regulation of cellular process
GO:0016043 0.73 cellular component organization
GO:0006996 0.63 organelle organization
GO:0048518 0.61 positive regulation of biological process
GO:0048522 0.58 positive regulation of cellular process
* GO:0007010 0.54 cytoskeleton organization
GO:0022607 0.53 cellular component assembly
* GO:0032185 0.47 septin cytoskeleton organization
* GO:0007017 0.47 microtubule-based process
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.98 intracellular part
GO:0044444 0.84 cytoplasmic part
GO:0043226 0.82 organelle
GO:0043229 0.80 intracellular organelle
GO:0044422 0.77 organelle part
GO:0044446 0.75 intracellular organelle part
GO:0043227 0.72 membrane-bounded organelle
GO:0005737 0.66 cytoplasm
GO:0043231 0.63 intracellular membrane-bounded organelle
GO:0016020 0.62 membrane
GO:0032991 0.59 protein-containing complex
* GO:0044430 0.55 cytoskeletal part
GO:0043232 0.54 intracellular non-membrane-bounded organelle
$$$$
Q96KV7-1 regulation of transcription by nucleic acid binding
==== MF ====
GO:1901363 0.59 heterocyclic compound binding
GO:0097159 0.59 organic cyclic compound binding
* GO:0140110 0.46 transcription regulator activity
GO:0003677 0.41 DNA binding
==== BP ====
GO:0009987 0.92 cellular process
GO:0065007 0.84 biological regulation
GO:0050789 0.82 regulation of biological process
GO:0050794 0.80 regulation of cellular process
GO:0019222 0.66 regulation of metabolic process
GO:0060255 0.64 regulation of macromolecule metabolic process
GO:0080090 0.63 regulation of primary metabolic process
GO:0031323 0.63 regulation of cellular metabolic process
GO:0051171 0.62 regulation of nitrogen compound metabolic process
* GO:0010468 0.60 regulation of gene expression
GO:0019219 0.59 regulation of nucleobase-containing compound metabolic process
GO:2000112 0.58 regulation of cellular macromolecule biosynthetic process
* GO:0051252 0.58 regulation of RNA metabolic process
GO:0071840 0.57 cellular component organization or biogenesis
* GO:0006355 0.57 regulation of transcription, DNA-templated
GO:0044237 0.56 cellular metabolic process
GO:0016043 0.54 cellular component organization
GO:0071704 0.53 organic substance metabolic process
GO:0044238 0.53 primary metabolic process
GO:0006807 0.53 nitrogen compound metabolic process
GO:0043170 0.52 macromolecule metabolic process
GO:0048519 0.46 negative regulation of biological process
GO:0048518 0.46 positive regulation of biological process
GO:0048523 0.44 negative regulation of cellular process
GO:0048522 0.44 positive regulation of cellular process
* GO:0006357 0.43 regulation of transcription by RNA polymerase II
==== CC ====
GO:0044424 1.00 intracellular part
GO:0043226 0.87 organelle
GO:0043229 0.86 intracellular organelle
GO:0043227 0.80 membrane-bounded organelle
GO:0043231 0.78 intracellular membrane-bounded organelle
GO:0044446 0.77 intracellular organelle part
GO:0032991 0.75 protein-containing complex
* GO:0005634 0.75 nucleus
* GO:0044428 0.56 nuclear part
$$$$
Q96M27-1 protein kinase A regulation
==== MF ====
* GO:0034237 0.65 protein kinase A regulatory subunit binding
==== BP ====
GO:0065007 0.94 biological regulation
GO:0050789 0.92 regulation of biological process
GO:0050794 0.91 regulation of cellular process
GO:0060255 0.82 regulation of macromolecule metabolic process
GO:0031323 0.81 regulation of cellular metabolic process
GO:0080090 0.80 regulation of primary metabolic process
GO:0051171 0.80 regulation of nitrogen compound metabolic process
GO:0048518 0.79 positive regulation of biological process
GO:0048522 0.77 positive regulation of cellular process
GO:0009987 0.77 cellular process
GO:0065008 0.74 regulation of biological quality
GO:0032501 0.74 multicellular organismal process
GO:0010604 0.73 positive regulation of macromolecule metabolic process
GO:0051173 0.72 positive regulation of nitrogen compound metabolic process
GO:0031325 0.72 positive regulation of cellular metabolic process
GO:0065009 0.71 regulation of molecular function
GO:0051246 0.71 regulation of protein metabolic process
GO:0032268 0.70 regulation of cellular protein metabolic process
GO:0050790 0.69 regulation of catalytic activity
GO:0031399 0.69 regulation of protein modification process
GO:0051247 0.68 positive regulation of protein metabolic process
GO:0044093 0.68 positive regulation of molecular function
GO:0019220 0.68 regulation of phosphate metabolic process
GO:0045859 0.67 regulation of protein kinase activity
GO:0042592 0.67 homeostatic process
* GO:0001934 0.67 positive regulation of protein phosphorylation
GO:0043085 0.66 positive regulation of catalytic activity
GO:0007389 0.66 pattern specification process
* GO:0034199 0.65 activation of protein kinase A activity
GO:0010669 0.65 epithelial structure maintenance
GO:0001757 0.65 somite specification
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.99 intracellular part
GO:0043226 0.92 organelle
GO:0043229 0.90 intracellular organelle
GO:0044444 0.89 cytoplasmic part
GO:0043227 0.88 membrane-bounded organelle
GO:0043231 0.86 intracellular membrane-bounded organelle
GO:0005737 0.72 cytoplasm
GO:0005794 0.59 Golgi apparatus
GO:0044422 0.54 organelle part
$$$$
Q96S16-1 histone demethylation
==== MF ====
GO:0016491 0.88 oxidoreductase activity
GO:0140096 0.87 catalytic activity, acting on a protein
GO:0051213 0.87 dioxygenase activity
* GO:0016705 0.87 oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen
* GO:0016706 0.79 2-oxoglutarate-dependent dioxygenase activity
* GO:0032452 0.75 histone demethylase activity
GO:0043167 0.66 ion binding
GO:1901363 0.65 heterocyclic compound binding
GO:0097159 0.65 organic cyclic compound binding
GO:0043169 0.64 cation binding
* GO:0003682 0.50 chromatin binding
==== BP ====
GO:0009987 0.99 cellular process
GO:0071704 0.97 organic substance metabolic process
GO:0044238 0.97 primary metabolic process
GO:0044237 0.97 cellular metabolic process
GO:1901564 0.96 organonitrogen compound metabolic process
GO:0043170 0.96 macromolecule metabolic process
GO:0043412 0.95 macromolecule modification
GO:0044260 0.92 cellular macromolecule metabolic process
GO:0019538 0.89 protein metabolic process
GO:0006464 0.88 cellular protein modification process
GO:0050789 0.84 regulation of biological process
GO:0050794 0.77 regulation of cellular process
GO:0019222 0.76 regulation of metabolic process
GO:0080090 0.75 regulation of primary metabolic process
GO:0060255 0.75 regulation of macromolecule metabolic process
GO:0031323 0.75 regulation of cellular metabolic process
GO:0051171 0.74 regulation of nitrogen compound metabolic process
GO:0019219 0.71 regulation of nucleobase-containing compound metabolic process
GO:0010468 0.71 regulation of gene expression
GO:0051252 0.70 regulation of RNA metabolic process
GO:0016043 0.67 cellular component organization
* GO:0018193 0.63 peptidyl-amino acid modification
* GO:0006325 0.63 chromatin organization
* GO:0016570 0.61 histone modification
* GO:0016577 0.52 histone demethylation
GO:0031326 0.50 regulation of cellular biosynthetic process
==== CC ====
GO:0044424 1.00 intracellular part
GO:0043226 0.95 organelle
GO:0043229 0.93 intracellular organelle
GO:0043227 0.93 membrane-bounded organelle
GO:0043231 0.90 intracellular membrane-bounded organelle
GO:0044446 0.86 intracellular organelle part
* GO:0005634 0.86 nucleus
* GO:0044428 0.79 nuclear part
GO:0044444 0.65 cytoplasmic part
* GO:0005654 0.61 nucleoplasm
GO:0032991 0.58 protein-containing complex
$$$$
Q9BZD6-1 serine-type endopeptidase
==== MF ====
GO:0140096 0.91 catalytic activity, acting on a protein
GO:0016787 0.90 hydrolase activity
* GO:0070011 0.89 peptidase activity, acting on L-amino acid peptides
* GO:0004175 0.84 endopeptidase activity
* GO:0008236 0.79 serine-type peptidase activity
* GO:0004252 0.74 serine-type endopeptidase activity
GO:0043167 0.53 ion binding
==== BP ====
GO:0065007 0.96 biological regulation
GO:0065008 0.90 regulation of biological quality
GO:0050789 0.87 regulation of biological process
GO:0050878 0.82 regulation of body fluid levels
GO:0009987 0.78 cellular process
GO:0048583 0.77 regulation of response to stimulus
GO:0051239 0.71 regulation of multicellular organismal process
GO:0032101 0.64 regulation of response to external stimulus
GO:0048519 0.63 negative regulation of biological process
GO:0008152 0.63 metabolic process
GO:0071704 0.62 organic substance metabolic process
GO:0050794 0.60 regulation of cellular process
GO:0044238 0.60 primary metabolic process
GO:0080134 0.59 regulation of response to stress
GO:0050818 0.59 regulation of coagulation
GO:0048518 0.58 positive regulation of biological process
GO:0006807 0.57 nitrogen compound metabolic process
GO:0043170 0.56 macromolecule metabolic process
* GO:0030193 0.56 regulation of blood coagulation
GO:1901564 0.55 organonitrogen compound metabolic process
GO:0051241 0.54 negative regulation of multicellular organismal process
GO:0048585 0.54 negative regulation of response to stimulus
GO:0032501 0.52 multicellular organismal process
GO:0019538 0.51 protein metabolic process
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.93 intracellular part
GO:0005737 0.74 cytoplasm
GO:0043226 0.71 organelle
GO:0043227 0.69 membrane-bounded organelle
GO:0044421 0.67 extracellular region part
GO:0005615 0.65 extracellular space
GO:0043229 0.64 intracellular organelle
GO:0043231 0.61 intracellular membrane-bounded organelle
$$$$
Q9BZH6-1
==== MF ====
GO:0097159 0.60 organic cyclic compound binding
GO:1901363 0.59 heterocyclic compound binding
==== BP ====
GO:0009987 0.97 cellular process
GO:0065007 0.85 biological regulation
GO:0050789 0.82 regulation of biological process
GO:0050794 0.80 regulation of cellular process
GO:0044237 0.75 cellular metabolic process
GO:0071704 0.73 organic substance metabolic process
GO:0044238 0.73 primary metabolic process
GO:0071840 0.72 cellular component organization or biogenesis
GO:0006807 0.72 nitrogen compound metabolic process
GO:0043170 0.71 macromolecule metabolic process
GO:0016043 0.69 cellular component organization
GO:0048518 0.63 positive regulation of biological process
GO:0019222 0.62 regulation of metabolic process
GO:0060255 0.61 regulation of macromolecule metabolic process
GO:0031323 0.61 regulation of cellular metabolic process
GO:0080090 0.60 regulation of primary metabolic process
GO:0051171 0.60 regulation of nitrogen compound metabolic process
GO:0048522 0.59 positive regulation of cellular process
GO:0050896 0.58 response to stimulus
GO:1901360 0.56 organic cyclic compound metabolic process
GO:0044260 0.56 cellular macromolecule metabolic process
GO:0034641 0.56 cellular nitrogen compound metabolic process
GO:0032502 0.56 developmental process
GO:0006725 0.56 cellular aromatic compound metabolic process
GO:0046483 0.55 heterocycle metabolic process
GO:0022607 0.55 cellular component assembly
GO:0090304 0.54 nucleic acid metabolic process
GO:0019219 0.53 regulation of nucleobase-containing compound metabolic process
GO:0048583 0.51 regulation of response to stimulus
GO:0009893 0.51 positive regulation of metabolic process
GO:0051173 0.50 positive regulation of nitrogen compound metabolic process
GO:0043933 0.50 protein-containing complex subunit organization
GO:0031325 0.50 positive regulation of cellular metabolic process
GO:0010604 0.50 positive regulation of macromolecule metabolic process
==== CC ====
GO:0044424 1.00 intracellular part
GO:0043229 0.97 intracellular organelle
GO:0032991 0.96 protein-containing complex
GO:0044446 0.95 intracellular organelle part
GO:0043227 0.89 membrane-bounded organelle
GO:0043231 0.88 intracellular membrane-bounded organelle
GO:0044444 0.87 cytoplasmic part
GO:0005737 0.82 cytoplasm
GO:0044428 0.72 nuclear part
GO:0005634 0.71 nucleus
GO:1902494 0.57 catalytic complex
GO:0044425 0.55 membrane part
GO:0005829 0.50 cytosol
$$$$
Q9C0D6-1 binding of cytoskeleton
==== MF ====
* GO:0008092 0.70 cytoskeletal protein binding
GO:0019899 0.48 enzyme binding
GO:0016787 0.41 hydrolase activity
==== BP ====
GO:0009987 0.95 cellular process
GO:0065007 0.82 biological regulation
GO:0050789 0.80 regulation of biological process
GO:0050794 0.75 regulation of cellular process
GO:0016043 0.69 cellular component organization
GO:0032502 0.54 developmental process
GO:0048518 0.48 positive regulation of biological process
GO:0051128 0.46 regulation of cellular component organization
GO:0065008 0.44 regulation of biological quality
GO:0006996 0.43 organelle organization
GO:0022607 0.42 cellular component assembly
GO:0048522 0.41 positive regulation of cellular process
GO:0006928 0.40 movement of cell or subcellular component
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.99 intracellular part
GO:0032991 0.87 protein-containing complex
GO:0044444 0.85 cytoplasmic part
GO:0043226 0.84 organelle
GO:0044422 0.83 organelle part
GO:0043229 0.83 intracellular organelle
GO:0044446 0.82 intracellular organelle part
GO:0043227 0.72 membrane-bounded organelle
GO:0005737 0.69 cytoplasm
GO:0043231 0.68 intracellular membrane-bounded organelle
GO:0016020 0.64 membrane
GO:0005634 0.53 nucleus
GO:0005829 0.48 cytosol
* GO:0044430 0.47 cytoskeletal part
GO:0043232 0.44 intracellular non-membrane-bounded organelle
GO:0044428 0.43 nuclear part
GO:0044425 0.40 membrane part
$$$$
Q9GZU8-1 hydrolase, probably hydrolase of protein
==== MF ====
GO:0043167 0.54 ion binding
* GO:0016787 0.53 hydrolase activity
* GO:0140096 0.48 catalytic activity, acting on a protein
==== BP ====
GO:0009987 0.88 cellular process
GO:0065007 0.82 biological regulation
GO:0050789 0.80 regulation of biological process
GO:0050794 0.79 regulation of cellular process
GO:0008152 0.70 metabolic process
GO:0071704 0.67 organic substance metabolic process
GO:0044238 0.67 primary metabolic process
GO:0006807 0.65 nitrogen compound metabolic process
GO:0043170 0.61 macromolecule metabolic process
GO:1901564 0.55 organonitrogen compound metabolic process
GO:0071840 0.53 cellular component organization or biogenesis
GO:0048519 0.52 negative regulation of biological process
GO:0016043 0.52 cellular component organization
GO:0048523 0.51 negative regulation of cellular process
GO:0044237 0.51 cellular metabolic process
* GO:0019538 0.50 protein metabolic process
==== CC ====
GO:0044464 0.99 cell part
GO:0044424 0.98 intracellular part
GO:0044422 0.73 organelle part
GO:0005576 0.69 extracellular region
GO:0044446 0.68 intracellular organelle part
GO:0043226 0.67 organelle
GO:0043229 0.66 intracellular organelle
GO:0044444 0.58 cytoplasmic part
GO:0043227 0.58 membrane-bounded organelle
GO:0043231 0.55 intracellular membrane-bounded organelle
GO:0016020 0.54 membrane
GO:0044425 0.51 membrane part
$$$$
Q9H5V9-1 mRNA splicing
==== MF ====
==== BP ====
* GO:0000398 1.00 mRNA splicing, via spliceosome
==== CC ====
GO:0044424 1.00 intracellular part
* GO:1902494 0.74 catalytic complex
* GO:0071013 0.70 catalytic step 2 spliceosome
$$$$
Q9H9L7-1 by binding to RNA polymerase, regulate expression of genes such as cytokines
==== MF ====
* GO:0019899 0.77 enzyme binding
==== BP ====
* GO:0045944 1.00 positive regulation of transcription by RNA polymerase II
GO:0050896 0.91 response to stimulus
GO:0009605 0.90 response to external stimulus
GO:0043207 0.89 response to external biotic stimulus
GO:0009987 0.85 cellular process
GO:1901700 0.71 response to oxygen-containing compound
GO:0051239 0.71 regulation of multicellular organismal process
GO:0010033 0.71 response to organic substance
GO:0051240 0.70 positive regulation of multicellular organismal process
GO:0033993 0.69 response to lipid
GO:0032496 0.68 response to lipopolysaccharide
* GO:0001819 0.68 positive regulation of cytokine production
* GO:0032755 0.67 positive regulation of interleukin-6 production
GO:0032502 0.65 developmental process
GO:0048856 0.61 anatomical structure development
==== CC ====
* GO:0005634 1.00 nucleus
GO:0032991 0.90 protein-containing complex
GO:0044422 0.76 organelle part
GO:0044446 0.75 intracellular organelle part
GO:0017053 0.65 transcriptional repressor complex
GO:0044428 0.58 nuclear part
GO:0044444 0.52 cytoplasmic part
$$$$
Q9NVP4-1
==== MF ====
==== BP ====
GO:0009987 0.93 cellular process
GO:0065007 0.85 biological regulation
GO:0050789 0.80 regulation of biological process
GO:0050794 0.79 regulation of cellular process
GO:0032502 0.51 developmental process
GO:0016043 0.50 cellular component organization
==== CC ====
GO:0044464 1.00 cell part
GO:0044424 0.98 intracellular part
GO:0043226 0.79 organelle
GO:0043229 0.78 intracellular organelle
GO:0005737 0.72 cytoplasm
GO:0043227 0.69 membrane-bounded organelle
GO:0044444 0.68 cytoplasmic part
GO:0043231 0.63 intracellular membrane-bounded organelle
GO:0044422 0.56 organelle part
GO:0044446 0.52 intracellular organelle part
$$$$