About COFACTOR
COFACTOR generates protein function annotation by searching query protein against three independent sequence, structure, and PPI function library.
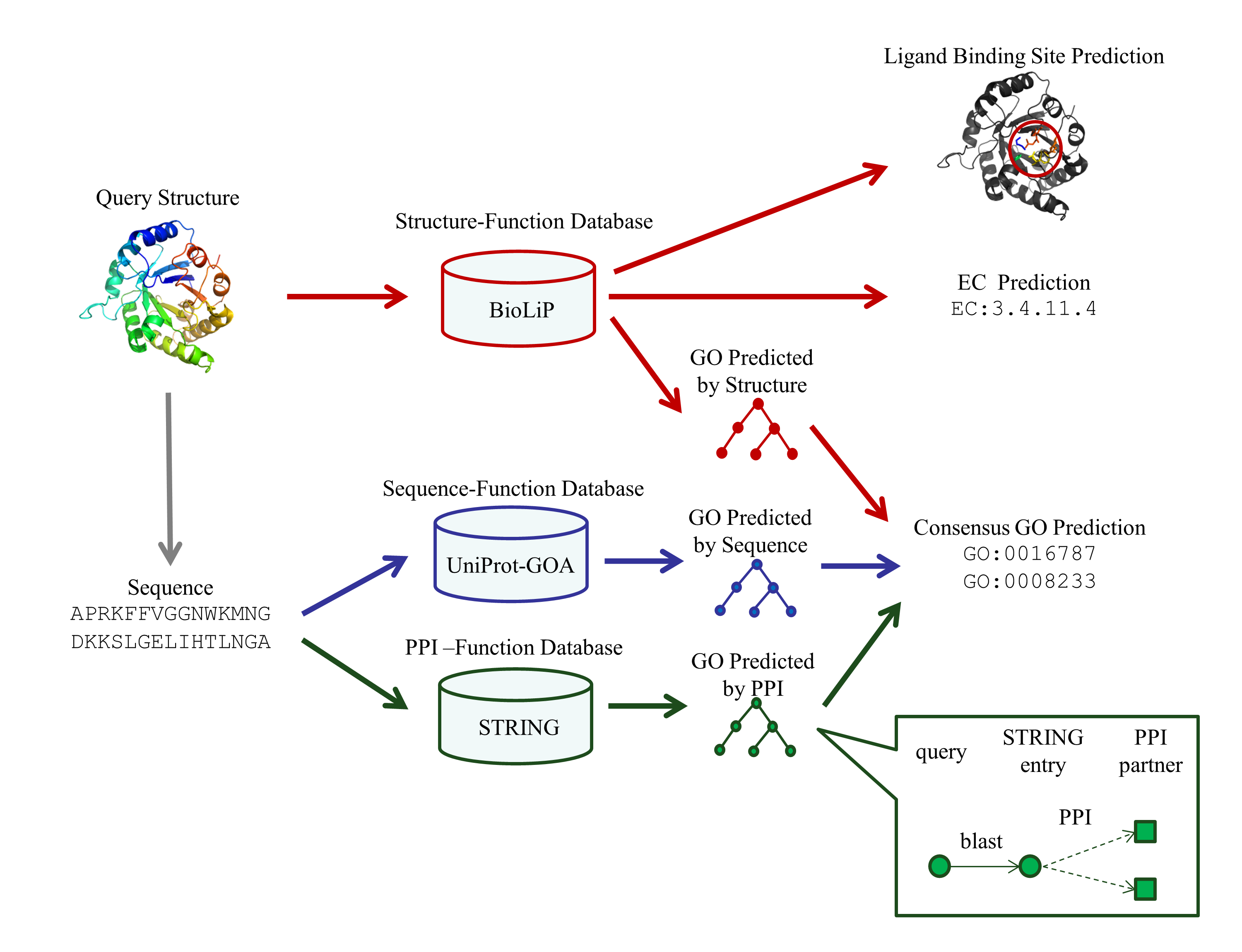
User Input
The user must upload a PDB formatted structure file of query protein. User's have the option to either copy the PDB file in the provided form or upload the file using the browse button. If the PDB file contains multiple protein chains and/or multiple models, first protein chain of the first model will be automatically selected by the program. Providing an e-mail address is recommended for recieving the prediction results.
Advanced Options.
Sequence of Query Protein:
By default, the input sequence for sequence- and PPI-based pipeline are extracted from structure file input by user. However, in many experimental structures, some residues of the full length protein are missing. In this case, the user is encouraged to provide the full length query protein sequence. Given user specified sequence, the COFACTOR structure-based pipeline will still use the structure file, while the sequence- and PPI-based pipelines will use the users specified sequence as input.
Exclude template: COFACTOR draws function insights from templates identified by searching the query against three funtion databases: BioLiP, UniProt-GOA, and STRING. If "remove templates sharing sequence identity 0.3 with target" was choosen, function prediction will not be generated from function templates that are highly homologous to target sequence. In general, excluding homologous templates will make structure prediction harder. So this option is only for benchmarking purposes.
Prediction results
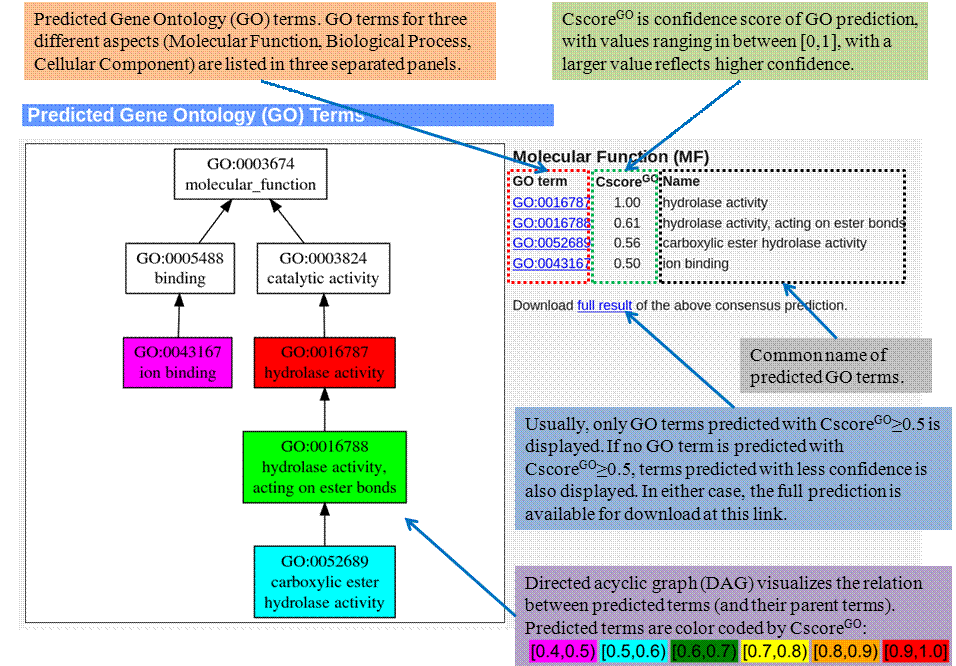
Gene Ontology (GO) terms
Predicted GO terms for Molecular Function (MF), Biological Process (BP), and Cellular Component (CC) are enlisted separated in this section, with graphic representation of relation between predicted terms.
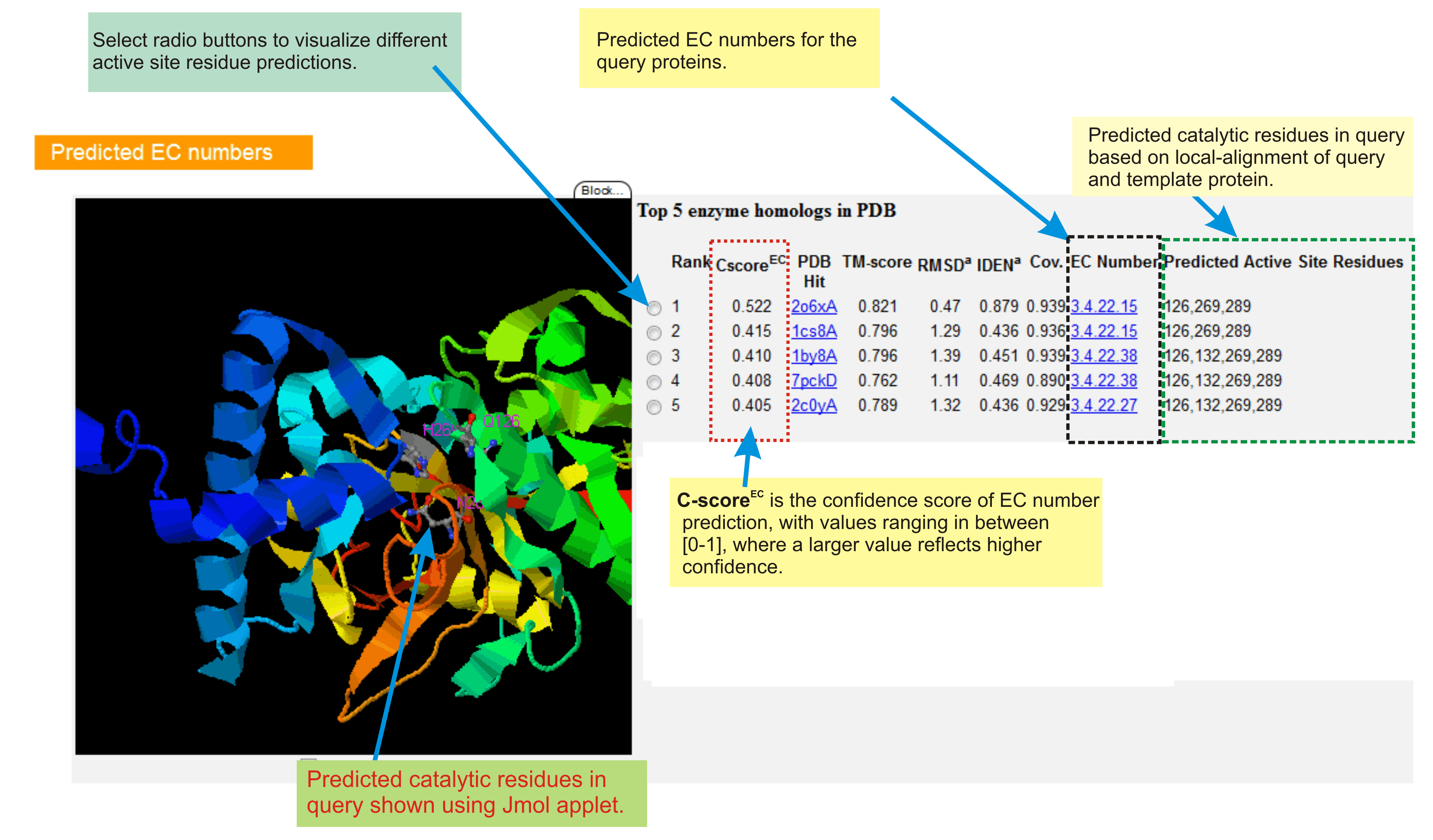
Enzyme Commission (EC) numbers
The section enlists top 5 enzyme homologs detected in PDB using COFACTOR global-local structure alignment procedure. Catalytic residues in query protein are also predicted based on active-site match of query and template proteins.
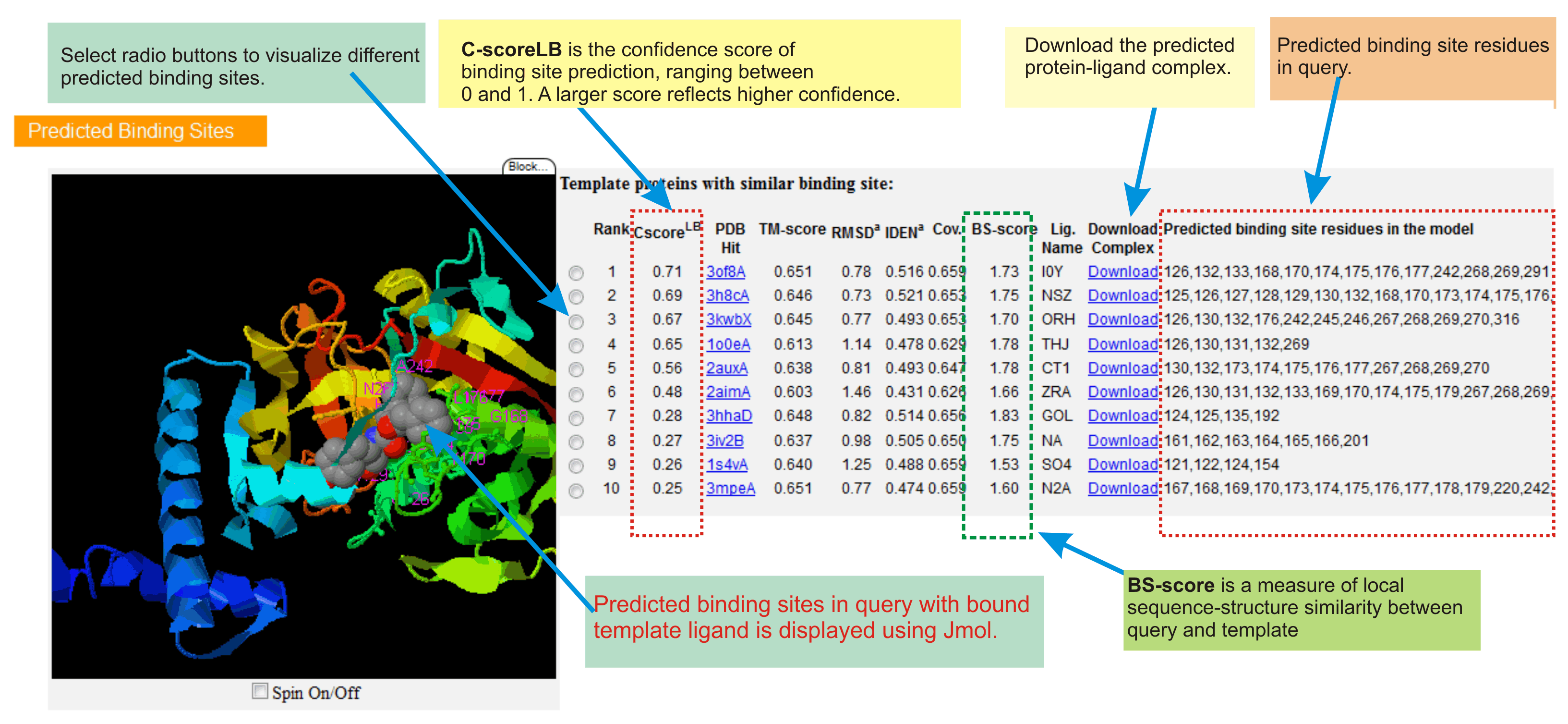
Protein-ligand binding site predictions
This section contains top 10 predicted ligand binding sites in query protein.
[an error occurred while processing this directive]




